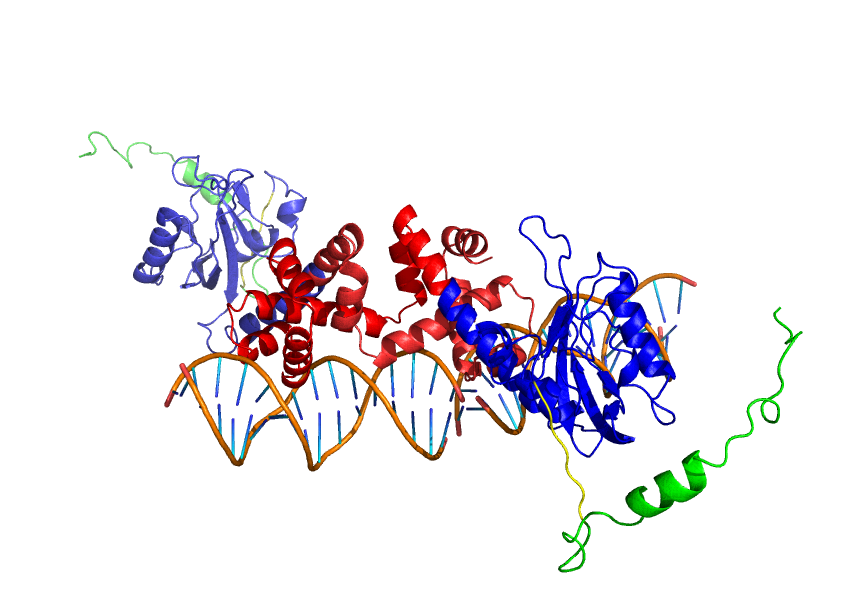
To better understand the possibilities of this project, we've modeled different aspects of the protein and the elements around it. Please select the subject you want to know more about:
Why and how we think the LovTAP-VP16 protein works?
What proportion of LovTAP-VP16 should we expect to be active depending on the lightning pattern?
Getting a rough idea of the expression levels of reporter protein we can expect.
Simulating the bioreactor lighting will give us an idea of the scalability of this type of production method.
We built our lighting device from scratch. Here are a few pictures of that process.
 "
"