|
|
| (30 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| - | <!-- *** What falls between these lines is the Alert Box! You can remove it from your pages once you have read and understood the alert *** -->
| + | {{:Team:EPF-Lausanne/Template/Header|project}} |
| | | | |
| - | <html>
| + | {{:Team:EPF-Lausanne/Template/SetTitle|Parts Submitted to the Registry}} |
| - | <div id="box" style="width: 700px; margin-left: 137px; padding: 5px; border: 3px solid #000; background-color: #fe2b33;">
| + | |
| - | <div id="template" style="text-align: center; font-weight: bold; font-size: large; color: #f6f6f6; padding: 5px;">
| + | |
| - | This is a template page. READ THESE INSTRUCTIONS.
| + | |
| - | </div>
| + | |
| - | <div id="instructions" style="text-align: center; font-weight: normal; font-size: small; color: #f6f6f6; padding: 5px;">
| + | |
| - | You are provided with this team page template with which to start the iGEM season. You may choose to personalize it to fit your team but keep the same "look." Or you may choose to take your team wiki to a different level and design your own wiki. You can find some examples <a href="https://2008.igem.org/Help:Template/Examples">HERE</a>.
| + | |
| - | </div>
| + | |
| - | <div id="warning" style="text-align: center; font-weight: bold; font-size: small; color: #f6f6f6; padding: 5px;">
| + | |
| - | You <strong>MUST</strong> have all of the pages listed in the menu below with the names specified. PLEASE keep all of your pages within your teams namespace.
| + | |
| - | </div>
| + | |
| - | </div>
| + | |
| - | </html>
| + | |
| | | | |
| - | <!-- *** End of the alert box *** -->
| + | ===Our parts=== |
| | | | |
| - | {| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center"
| + | We managed to submit 3 new and characterized parts that will give other teams the possibility to reproduce our experiments with both the switches we tested. |
| - | !align="center"|[[Team:EPF-Lausanne|Home]]
| + | |
| - | !align="center"|[[Team:EPF-Lausanne/Team|Team]]
| + | |
| - | !align="center"|[https://igem.org/Team.cgi?year=2012&team_name=EPF-Lausanne Official Team Profile]
| + | |
| - | !align="center"|[[Team:EPF-Lausanne/Project|Project]]
| + | |
| - | !align="center"|[[Team:EPF-Lausanne/Parts|Parts Submitted to the Registry]]
| + | |
| - | !align="center"|[[Team:EPF-Lausanne/Modeling|Modeling]]
| + | |
| - | !align="center"|[[Team:EPF-Lausanne/Notebook|Notebook]]
| + | |
| - | !align="center"|[[Team:EPF-Lausanne/Safety|Safety]]
| + | |
| - | !align="center"|[[Team:EPF-Lausanne/Attributions|Attributions]]
| + | |
| - | |}
| + | |
| | | | |
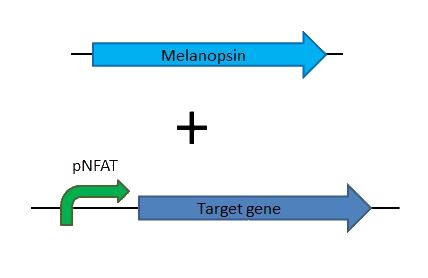
| | + | The [http://partsregistry.org/Part:BBa_K838002 melanopsin coding gene] will allow to use the more complex switch, by just cloning melanopsin into a mammalian vector and cloning a readout gene (fluorescent protein, quantitatively measurable protein) into the commercially available mammalian vector [http://www.promega.com/resources/protocols/product-information-sheets/a/pgl430-vector-protocol/ pGL4.30] containing pNFAT, the promoter activated by presence of NFAT which is the outcome of the calcium channel mediated pathway. |
| | | | |
| | + | We also submitted our favorite parts, which are a mammalian [http://partsregistry.org/Part:BBa_K838000 LovTAP-VP16] and a [http://partsregistry.org/Part:BBa_K838001 composite readout system] build to respond to LovTAP-VP16 activation. |
| | + | Both parts only need cloning into a mammalian vector to be ready for transfection. |
| | | | |
| - | An important aspect of the iGEM competition is the use and creation of standard biological parts. Each team will make new parts during iGEM and will place them in the [http://partsregistry.org Registry of Standard Biological Parts]. The iGEM software provides an easy way to present the parts your team has created . The "groupparts" tag will generate a table with all of the parts that your team adds to your team sandbox. Note that if you want to document a part you need to document it on the [http://partsregistry.org Registry], not on your team wiki.
| |
| | | | |
| - | Remember that the goal of proper part documentation is to describe and define a part such that it can be used without a need to refer to the primary literature. The next iGEM team should be able to read your documentation and be able to use the part successfully. Also, you should provide proper references to acknowledge previous authors and to provide for users who wish to know more.
| + | <groupparts>iGEM012 EPF-Lausanne</groupparts> |
| | | | |
| | + | ===How to use=== |
| | | | |
| - | <groupparts>iGEM012 EPF-Lausanne</groupparts>
| + | '''1) Cloning:''' |
| | + | |
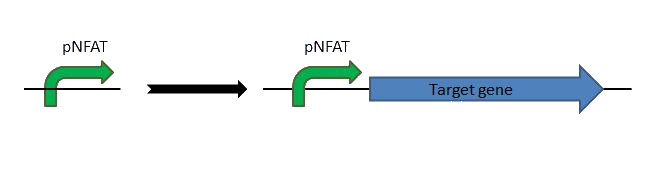
| | + | First clone the part into a mammalian expression vector such as [http://products.invitrogen.com/ivgn/product/V79020?ICID=search-product pcDNA3.1(+)] or [http://products.invitrogen.com/ivgn/product/V04450 pCEP4]. These are the two main expression vectors we used. |
| | + | To use the melanopsin pathway, you also need to clone any gene you want to express into the mammalian vector [http://www.promega.com/resources/protocols/product-information-sheets/a/pgl430-vector-protocol/ pGL4.30]. |
| | + | |
| | + | [[File:Team-EPF-Lausanne_pNFAT.png|500px]] |
| | + | |
| | + | '''2) Choose your switch:''' |
| | + | |
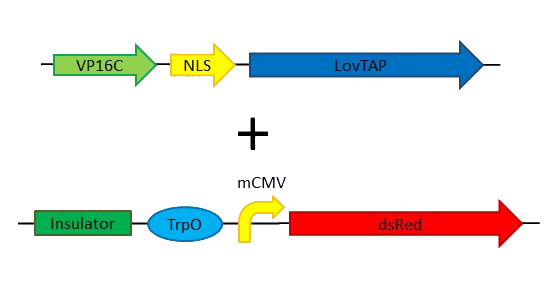
| | + | *Simple switch = LovTAP-VP16 + LovTAP readout |
| | + | [[File:Team_EPF_Lausanne_simpleswitch.png|500px]] |
| | + | *Complex switch = Melanopsin + pNFAT-target gene |
| | + | [[File:Team_EPF_Lausanne_complexswitch.png|400px]] |
| | + | |
| | + | '''3) Transfection:''' |
| | + | |
| | + | Co-transfect the combination of photoreceptive protein and readout construct of your choice in mammalian cells! |
| | + | |
| | + | |
| | + | [[File:Team-EPF-Lausanne-sheep.png|frameless|right|300px]] |
| | + | |
| | + | |
| | + | {{:Team:EPF-Lausanne/Template/Footer}} |
 "
"