Team:USP-UNESP-Brazil/Plasmid Plug n Play/Results
From 2012.igem.org
Andresochoa (Talk | contribs) |
|||
| (13 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
This experiment was made to test if the loxP and the lox66 were working properly, it also allowed us to measure the Cre concentration needed for an ''in vitro'' recombination reaction. The particularity of lox66 is that it has an altered sequence at the end of it's left arm when compared to loxP (natural recombination site from P1 Bacteriophage). This sequence variation reduces affinity of the Cre recombinase for the arm. | This experiment was made to test if the loxP and the lox66 were working properly, it also allowed us to measure the Cre concentration needed for an ''in vitro'' recombination reaction. The particularity of lox66 is that it has an altered sequence at the end of it's left arm when compared to loxP (natural recombination site from P1 Bacteriophage). This sequence variation reduces affinity of the Cre recombinase for the arm. | ||
| - | Based on the report made by the igem2010 UT-Tokyo team and some papers | + | Based on the report made by the igem2010 UT-Tokyo team and some papers [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC137435/], the lox66 (BBa_I718016) from the registry was wrong, it had a gg instead a cg in its left arm. This part was corrected by the iGEM11_Tokyo_Tech team (BBa_K649206) and by the iGEM11_WITS_CSIR_SA team (BBa_K537019), but no DNA was available in the registry. Anyway, we needed to synthesized it and test it as part of our PCR-primers, we used the proper sequence described by [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC137435/]. |
| + | |||
We designed two primers, one containing the lox66 and one containing the loxP sequences, these primers amplified the ORF from the kanamycin resistance gene, flanked upstream by the loxP and downstream by the lox66, using PCR. These sites should be recognized by the Cre recombinase (from NEB company), which could circularized our linear PCR product. This is important because we don't want it to be degraded when inserted in the bacteria. This ''In vitro'' assay was a test for a posterior ''In vivo'' assay, where we expect that this process happens inside the ''E. coli'' using a Cre recombinase enzyme expressed by the same bacteria. | We designed two primers, one containing the lox66 and one containing the loxP sequences, these primers amplified the ORF from the kanamycin resistance gene, flanked upstream by the loxP and downstream by the lox66, using PCR. These sites should be recognized by the Cre recombinase (from NEB company), which could circularized our linear PCR product. This is important because we don't want it to be degraded when inserted in the bacteria. This ''In vitro'' assay was a test for a posterior ''In vivo'' assay, where we expect that this process happens inside the ''E. coli'' using a Cre recombinase enzyme expressed by the same bacteria. | ||
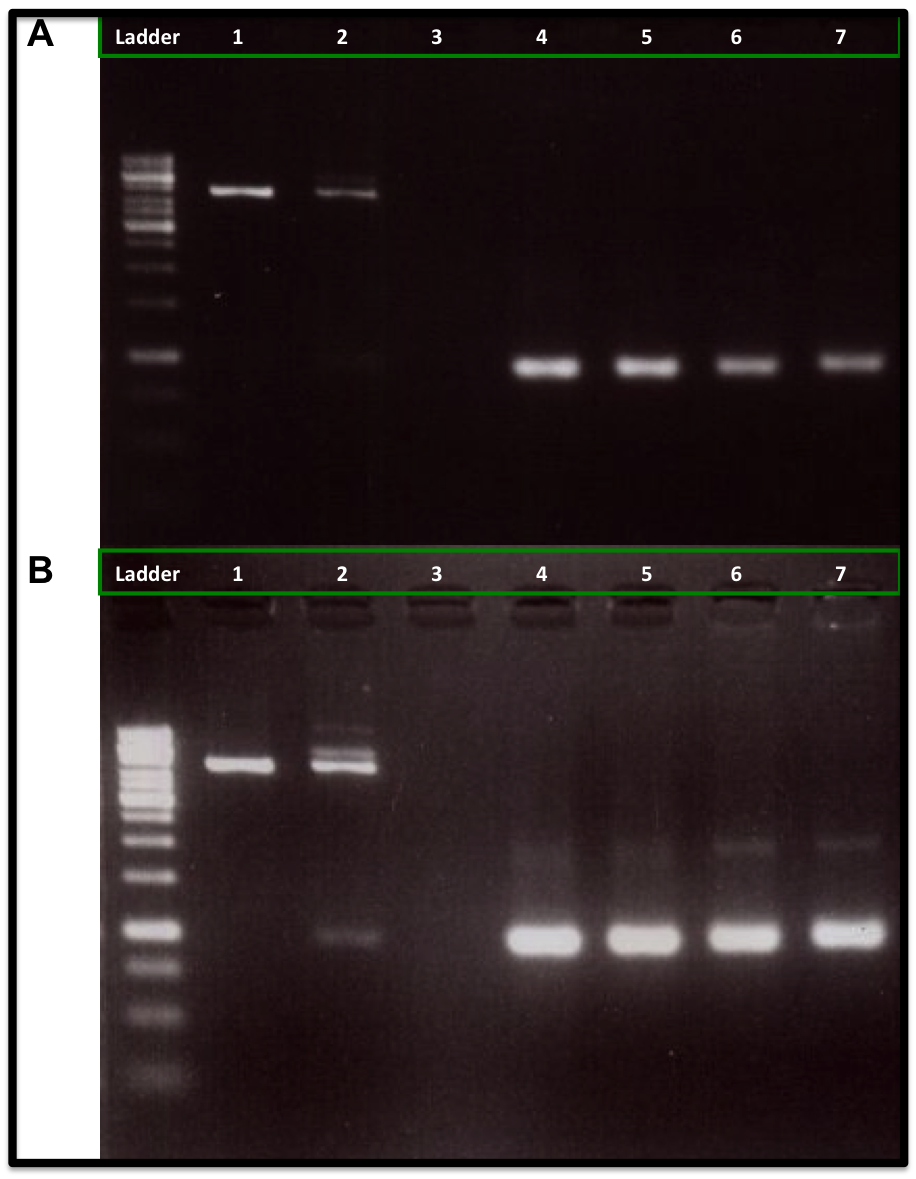
| - | Our experiment showed that 5U and 10U of Cre recombinase | + | Our experiment showed that 5U and 10U of Cre recombinase reduced the linear DNA (Kanamycin resistance gene flanked with loxP and lox66) amount when compare to 1U of Cre recombinase and to the control DNA (No Cre recombinase added), as is showed in the figure A. It was also observed an increase of the DNA plasmid form (upper band at 2 Kb), as is showed in figure B. We also used a control DNA substrate supplied in the NEB recombinase kit. The same amount of PCR-product was applied to each lane (260 ng). |
The conclusion was that we can use this loxP-lox66 mechanism in our design and we will need at least 5U of Cre recombinase for any ''in vitro'' experiment. | The conclusion was that we can use this loxP-lox66 mechanism in our design and we will need at least 5U of Cre recombinase for any ''in vitro'' experiment. | ||
{{:Team:USP-UNESP-Brazil/Templates/RImage | image=Reulst1.png | caption= 1kb ladder, 1) Control substrate from the NEB kit without Cre recombinase, 2) Control substrate from the NEB kit with 1U Cre, 3) Empty, 4) Kanamycin resistance gene without Cre recombinase, 5) Kanamycin resistance gene with 1U Cre, 6) Kanamycin resistance gene with 5U Cre, 7) Kanamycin resistance gene with 10U Cre. | size=600px }} | {{:Team:USP-UNESP-Brazil/Templates/RImage | image=Reulst1.png | caption= 1kb ladder, 1) Control substrate from the NEB kit without Cre recombinase, 2) Control substrate from the NEB kit with 1U Cre, 3) Empty, 4) Kanamycin resistance gene without Cre recombinase, 5) Kanamycin resistance gene with 1U Cre, 6) Kanamycin resistance gene with 5U Cre, 7) Kanamycin resistance gene with 10U Cre. | size=600px }} | ||
| + | |||
| Line 23: | Line 25: | ||
For this experiment we needed a bacteria lineage with the Plug&Play prototype plasmid. So we transformed electrocompetent ''E. coli'' (Bl21(DE3)) cells with 1 uL (80 ng) of the Plug&Play prototype. Then, we prepared electrocompetent cells using this transformed bacteria, producing a Plug&Play Machine ready to receive the kanamycin resistance gene. | For this experiment we needed a bacteria lineage with the Plug&Play prototype plasmid. So we transformed electrocompetent ''E. coli'' (Bl21(DE3)) cells with 1 uL (80 ng) of the Plug&Play prototype. Then, we prepared electrocompetent cells using this transformed bacteria, producing a Plug&Play Machine ready to receive the kanamycin resistance gene. | ||
| - | We transformed the electrocompetent Plug&Play Machine cells with a gradient of our purified PCR-product (kanamycin resistance gene flanked by a loxP and a lox66): 10 ng, 100 ng and 1000 ng. | + | We transformed the electrocompetent Plug&Play Machine cells with a gradient of our purified PCR-product (kanamycin resistance gene flanked by a loxP and a lox66): 10 ng, 100 ng and 1000 ng. These concentrations were chosen using the mathematical model that the group already had developed. After transformation, the cells (50 ul) were left for 40 min in 1 mL LB medium without antibiotic, for recovering from the transformation stress. |
| - | The LB medium (1 mL) | + | The LB medium (1 mL) where the bacteria were recovering was equally divided in three LB solid plates. One plate had ampicillin (100 ug/mL) and IPTG (1 mM), the second had kanamycin (50 ug/mL) and IPTG (1 mM) and the last one had both antibiotics (ampicillin (100 ug/mL) and kanamycin (50 ug/mL)) and IPTG (1 mM). The plates were left in incubation for 15 h at 37°C. As a control, the pET15b (25 ng) commercial plasmid was used for an independent transformation of the ''E. coli'' (Bl21(DE3)) lineage and plated in the same three kind of plates. The pET15b plasmid has a gene that confers resistance to ampicillin. |
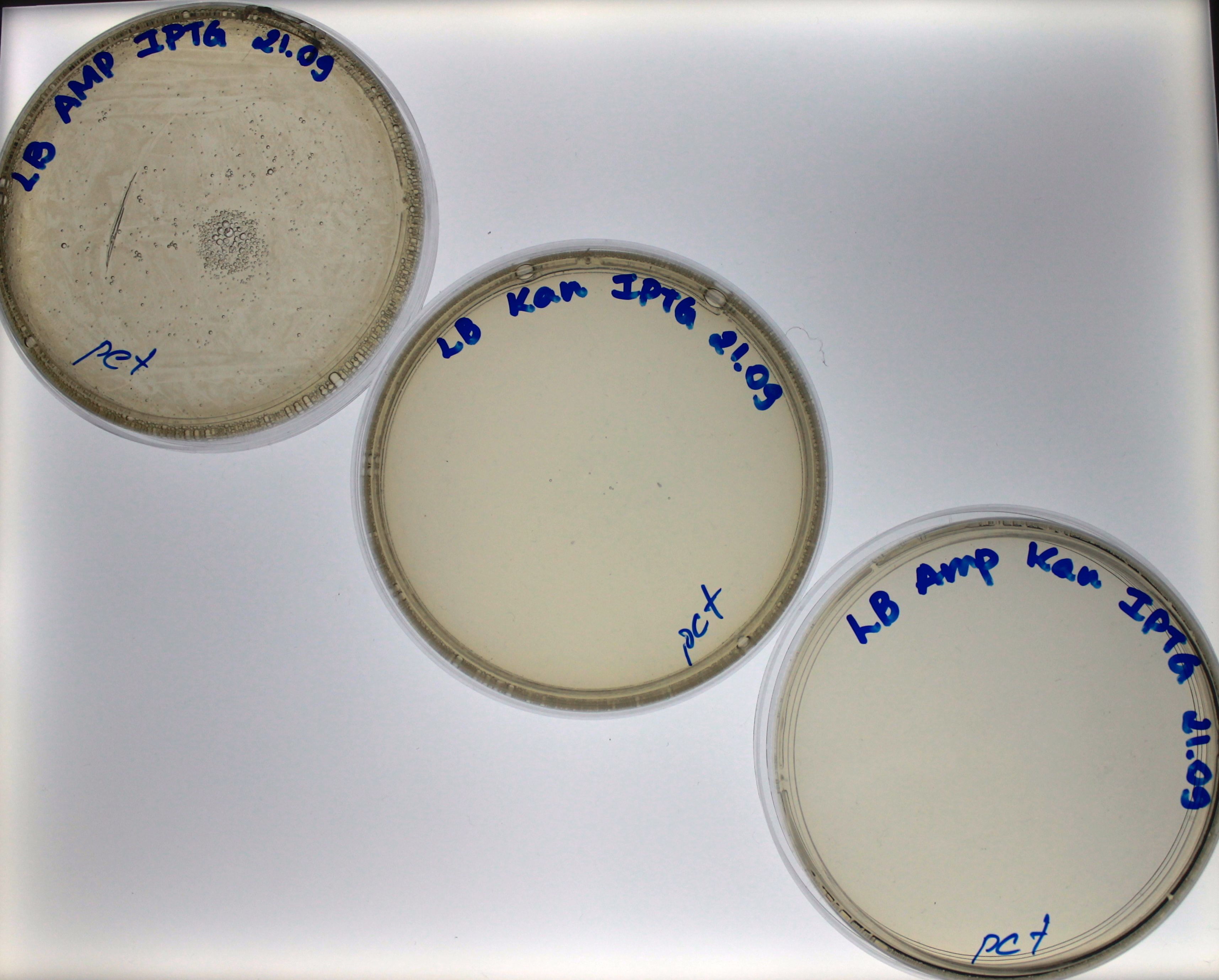
Our control worked as expected, the bacteria grew in the ampicillin plate but no in the kanamycin plate nor the plate with both antibiotics (Fig. 1). For the bacteria transformed with 10 ng of PCR-product, we found growth just in the ampicillin plate, which means that the bacteria had the Plug&Play plasmid, but no recombination had happened (Fig. 2). | Our control worked as expected, the bacteria grew in the ampicillin plate but no in the kanamycin plate nor the plate with both antibiotics (Fig. 1). For the bacteria transformed with 10 ng of PCR-product, we found growth just in the ampicillin plate, which means that the bacteria had the Plug&Play plasmid, but no recombination had happened (Fig. 2). | ||
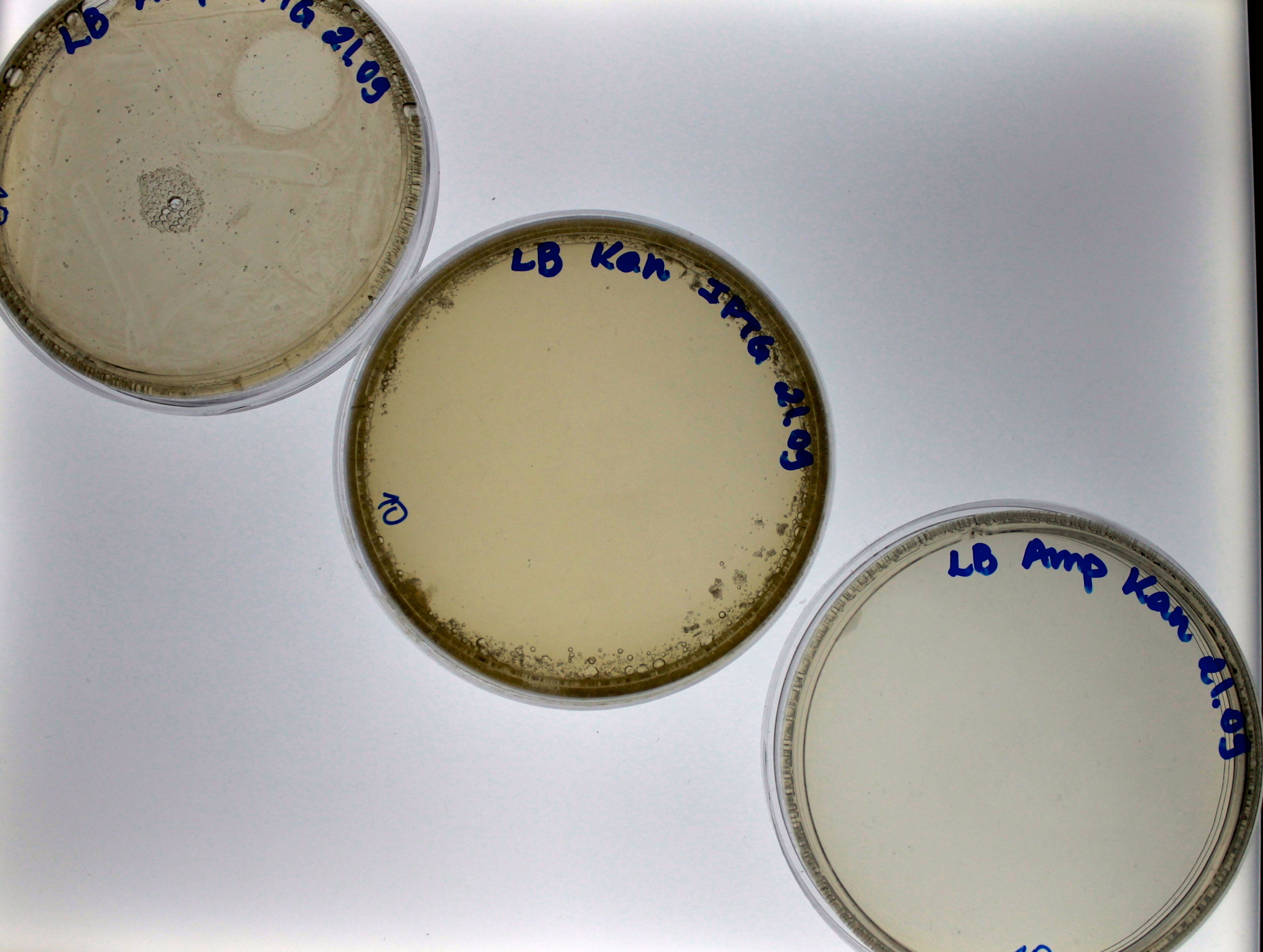
| - | For the bacteria transformed with 100 ng and 1000 ng PCR-products we found colonies in the three plates. | + | For the bacteria transformed with 100 ng and 1000 ng of PCR-products we found colonies in the three plates. Find colonies in the plate with both antibiotics (Fig. 3 and Fig. 4) strongly suggest that the bacteria had a Plug&Play plasmid where occurred the PCR-product recombination into the lox71 site. We counted the number of colonies in both plates: 36 colonies grew in the plate of bacteria transformed with 10 ng of PCR-product and 611 for the plate of bacteria transformed with 100 ng of PCR-product. |
| + | |||
| + | The kanamycin gene that we used cant express or replicate without being in the plasmid, it has no promoter or replication site, we cloned just the ORF of the gene. This gene will be expressed as long as IPTG is present in the culture medium. | ||
| + | |||
| + | |||
| Line 41: | Line 47: | ||
| - | {{:Team:USP-UNESP-Brazil/Templates/RImage | image=100exp4.png | caption= Fig. 3. LB plate (ampicillin plus kanamycin), used for the growth of Plug&Play Machine cells transformed with 100 ng of PCR purified product | size=500px }} | + | {{:Team:USP-UNESP-Brazil/Templates/RImage | image=100exp4.png | caption= Fig. 3. LB plate (ampicillin plus kanamycin), used for the growth of Plug&Play Machine cells transformed with 100 ng of PCR purified product. | size=500px }} |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | {{:Team:USP-UNESP-Brazil/Templates/RImage | image=Resultfinal1000.png | caption= Fig. 4.LB plate (ampicillin plus kanamycin), used for the growth of Plug&Play Machine cells transformed with 1000 ng of PCR purified product. | size=500px }} | ||
| Line 49: | Line 65: | ||
| + | <h1 id="''Conclusions'' ">''Conclusions'' </h1> | ||
| + | Our results suggest that this technology is possible, that the recombination system is able to circularize the PCR-product as soon as it enters in the cell, and before it is degraded by the cellular nucleases. This was also predicted in the results obtained from the mathematical model. | ||
| - | + | Based on the report made by the igem2010 UT-Tokyo team and some papers [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC137435/], the lox71 (BBa_I718017) from the registry was wrong, it had a cg instead a gc in the 8bp spacer sequence between the arms. Apparently, this part was corrected by the iGEM11_WITS_CSIR_SA team (BBa_K537020), but the sequence of this group had the same error. The corrected part from iGEM11_Tokyo_Tech (BBa_K649205) was right but no DNA was available in the registry. So we decided to synthesized it and test it, using the proper sequence described in [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC137435/]. We submitted the correct DNA to the Registry and our results suggest that the lox71 submitted by our group is working. | |
| + | This was the first test of one of the six prototypes, we want to test the others in order to find out which is the best structure for developing a complete series of recombination plasmids that use this mechanism. As a proof of concept we developed a plasmid for expression in ''E. coli'', but this methodology could be use for developing plasmids for expression in another systems, like plants and yeast. | ||
| + | This technology could help in the screening/development of candidate genes in a high-throughput manner for creating new standard biological parts, making faster and easier to produce tools for hacking biosystems. | ||
{{:Team:USP-UNESP-Brazil/Templates/Foot}} | {{:Team:USP-UNESP-Brazil/Templates/Foot}} | ||
Latest revision as of 03:51, 27 September 2012
 Introduction
Introduction Project Overview
Project Overview Plasmid Plug&Play
Plasmid Plug&Play Associative Memory
Associative MemoryNetwork
 Extras
ExtrasIn vitro assay
This experiment was made to test if the loxP and the lox66 were working properly, it also allowed us to measure the Cre concentration needed for an in vitro recombination reaction. The particularity of lox66 is that it has an altered sequence at the end of it's left arm when compared to loxP (natural recombination site from P1 Bacteriophage). This sequence variation reduces affinity of the Cre recombinase for the arm.
Based on the report made by the igem2010 UT-Tokyo team and some papers [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC137435/], the lox66 (BBa_I718016) from the registry was wrong, it had a gg instead a cg in its left arm. This part was corrected by the iGEM11_Tokyo_Tech team (BBa_K649206) and by the iGEM11_WITS_CSIR_SA team (BBa_K537019), but no DNA was available in the registry. Anyway, we needed to synthesized it and test it as part of our PCR-primers, we used the proper sequence described by [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC137435/].
We designed two primers, one containing the lox66 and one containing the loxP sequences, these primers amplified the ORF from the kanamycin resistance gene, flanked upstream by the loxP and downstream by the lox66, using PCR. These sites should be recognized by the Cre recombinase (from NEB company), which could circularized our linear PCR product. This is important because we don't want it to be degraded when inserted in the bacteria. This In vitro assay was a test for a posterior In vivo assay, where we expect that this process happens inside the E. coli using a Cre recombinase enzyme expressed by the same bacteria.
Our experiment showed that 5U and 10U of Cre recombinase reduced the linear DNA (Kanamycin resistance gene flanked with loxP and lox66) amount when compare to 1U of Cre recombinase and to the control DNA (No Cre recombinase added), as is showed in the figure A. It was also observed an increase of the DNA plasmid form (upper band at 2 Kb), as is showed in figure B. We also used a control DNA substrate supplied in the NEB recombinase kit. The same amount of PCR-product was applied to each lane (260 ng).
The conclusion was that we can use this loxP-lox66 mechanism in our design and we will need at least 5U of Cre recombinase for any in vitro experiment.
In vivo assay
We proved that we can circularize a fragment of DNA (kanamycin resistance gene) flanked by a loxP and a lox66 sites in vitro, so we decided to test one of the Plug&Play prototypes (T7-lox71-Cre-pSB4A5). This plasmid produces a low copy number and has a gene that confers resistance to ampicillin. The Cre recombinase is under the control of the T7 promoter, the target gene (in this case the kanamycin resistance gene) will be inserted in the lox71 site upstream the Cre recombinase gene. When the gene is inserted it will be controlled by the same T7 promoter. This plasmid is one of the six prototypes we are developing, the comparison of these plasmids will identify the best system for the developing of this technology.
For this experiment we needed a bacteria lineage with the Plug&Play prototype plasmid. So we transformed electrocompetent E. coli (Bl21(DE3)) cells with 1 uL (80 ng) of the Plug&Play prototype. Then, we prepared electrocompetent cells using this transformed bacteria, producing a Plug&Play Machine ready to receive the kanamycin resistance gene.
We transformed the electrocompetent Plug&Play Machine cells with a gradient of our purified PCR-product (kanamycin resistance gene flanked by a loxP and a lox66): 10 ng, 100 ng and 1000 ng. These concentrations were chosen using the mathematical model that the group already had developed. After transformation, the cells (50 ul) were left for 40 min in 1 mL LB medium without antibiotic, for recovering from the transformation stress.
The LB medium (1 mL) where the bacteria were recovering was equally divided in three LB solid plates. One plate had ampicillin (100 ug/mL) and IPTG (1 mM), the second had kanamycin (50 ug/mL) and IPTG (1 mM) and the last one had both antibiotics (ampicillin (100 ug/mL) and kanamycin (50 ug/mL)) and IPTG (1 mM). The plates were left in incubation for 15 h at 37°C. As a control, the pET15b (25 ng) commercial plasmid was used for an independent transformation of the E. coli (Bl21(DE3)) lineage and plated in the same three kind of plates. The pET15b plasmid has a gene that confers resistance to ampicillin.
Our control worked as expected, the bacteria grew in the ampicillin plate but no in the kanamycin plate nor the plate with both antibiotics (Fig. 1). For the bacteria transformed with 10 ng of PCR-product, we found growth just in the ampicillin plate, which means that the bacteria had the Plug&Play plasmid, but no recombination had happened (Fig. 2).
For the bacteria transformed with 100 ng and 1000 ng of PCR-products we found colonies in the three plates. Find colonies in the plate with both antibiotics (Fig. 3 and Fig. 4) strongly suggest that the bacteria had a Plug&Play plasmid where occurred the PCR-product recombination into the lox71 site. We counted the number of colonies in both plates: 36 colonies grew in the plate of bacteria transformed with 10 ng of PCR-product and 611 for the plate of bacteria transformed with 100 ng of PCR-product.
The kanamycin gene that we used cant express or replicate without being in the plasmid, it has no promoter or replication site, we cloned just the ORF of the gene. This gene will be expressed as long as IPTG is present in the culture medium.
Conclusions
Our results suggest that this technology is possible, that the recombination system is able to circularize the PCR-product as soon as it enters in the cell, and before it is degraded by the cellular nucleases. This was also predicted in the results obtained from the mathematical model.
Based on the report made by the igem2010 UT-Tokyo team and some papers [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC137435/], the lox71 (BBa_I718017) from the registry was wrong, it had a cg instead a gc in the 8bp spacer sequence between the arms. Apparently, this part was corrected by the iGEM11_WITS_CSIR_SA team (BBa_K537020), but the sequence of this group had the same error. The corrected part from iGEM11_Tokyo_Tech (BBa_K649205) was right but no DNA was available in the registry. So we decided to synthesized it and test it, using the proper sequence described in [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC137435/]. We submitted the correct DNA to the Registry and our results suggest that the lox71 submitted by our group is working.
This was the first test of one of the six prototypes, we want to test the others in order to find out which is the best structure for developing a complete series of recombination plasmids that use this mechanism. As a proof of concept we developed a plasmid for expression in E. coli, but this methodology could be use for developing plasmids for expression in another systems, like plants and yeast.
This technology could help in the screening/development of candidate genes in a high-throughput manner for creating new standard biological parts, making faster and easier to produce tools for hacking biosystems.
 "
"