Team:Colombia/Project/Basics
From 2012.igem.org
(→The Basics) |
Sylvita1015 (Talk | contribs) |
||
| (35 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{https://2012.igem.org/User:Tabima}} | {{https://2012.igem.org/User:Tabima}} | ||
| + | |||
| + | <div class="right_box"> | ||
='''The Basics'''= | ='''The Basics'''= | ||
| - | [[File: | + | [[File:basics.jpg|650px|center|Project design summary]] |
| + | |||
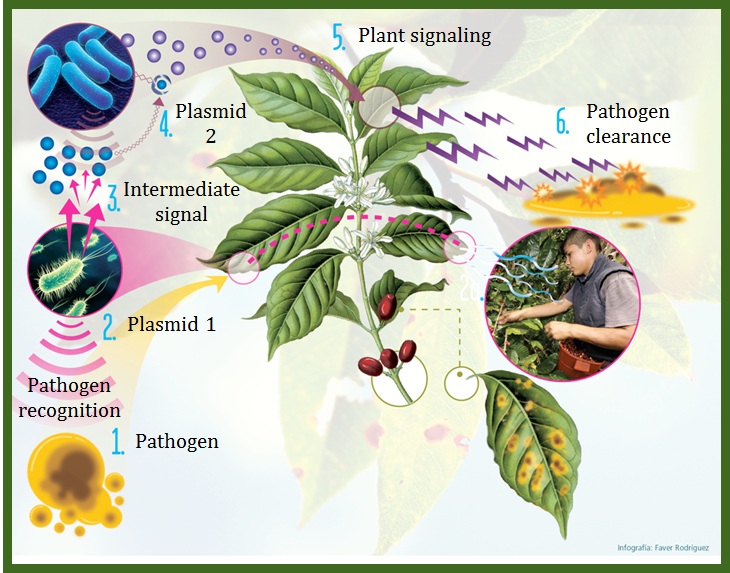
| + | Our objective is to generate a genetically-modified bacteria "detect and alert" system, particularly as a defense aid for crop plantations against plant pathogens (1). Bacteria are built (2,4) in a way that they can ''detect'' pathogen associated molecules, amplify this initial signal (3), and finally alert the plant (5) by stimulating an early hypersensitive response against infection (6). We have also included a way for the bacteria to control their own population density using a [http://en.wikipedia.org/wiki/Toxin-antitoxin_system Toxin-Antitoxin module] (TA). Using the bacteria ''Escherichia coli'' K12, the system will use two differently-modified plasmids. The first one will detect the designated pathogen associeted molecule and weigh such input for intensity and duration (2). If significant, it will also produce a signal molecule to engage the response of the second plasmid (3). Upon exposure to enough concentration of the molecule, the second plasmid will produce a systemic acquired response triggering hormone (salicylic acid) to activate the plant defenses (Loake & Grant, 2007) (5), both attacking the pathogen (6) as well as controlling the bacterial population growth. We strongly believe this biocontrol method may prove to become a powerful tool for farmers and crop workers everywhere. In addition, our constructed "detect and alert" biobricks could be useful to plentiful synthetic biology experiments and future iGEM teams. Finally, want to control against horizontal gene transfer and plasmid curing with using 2 different TA systems. | ||
| + | |||
| + | We're currently implementing [https://2012.igem.org/Team:Colombia/Project/Experiments/Our_Design our design ]for the control of ''Hemileia vastatrix'' [https://2012.igem.org/Team:Colombia/Project/Problem#Coffee_rust_.28Hemileia_vastatrix.29 coffee rust] by recognizing fungal cell wall chitin ([http://en.wikipedia.org/wiki/PAMP PAMP]), as well as the recognition of the ''Ralstonia solanacearum'' 3-hydroxypalmitic acid methyl ester (3-OH-PAME, a [http://en.wikipedia.org/wiki/Quorum_sensing quorum sensing] molecule [[References]] (Flavier ''et al.'', 1997)) against [https://2012.igem.org/Team:Colombia/Project/Problem#Bacterial_Wilt_.28Ralstonia_solanacearum.29 bacterial wilt]. Since the latter is a soil acquired disease, we are also evaluating the possibility of using [https://2012.igem.org/Team:Colombia/Project/Experiments/Pseudomonas ''Pseudomonas fluorescens''] instead of ''E. coli'' as a chassis. | ||
| + | |||
| + | =='''References'''== | ||
| - | + | #Flavier, A. B., Clough, S. J., Schell, M. A. and Denny, T. P. (1997), Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Molecular Microbiology, 26: 251–259. [http://onlinelibrary.wiley.com/doi/10.1046/j.1365-2958.1997.5661945.x/pdf doi:10.1046/j.1365-2958.1997.5661945.x] | |
| + | #Loake, G., & Grant, M. (2007). Salicylic acid in plant defence--the players and protagonists. Current opinion in plant biology, 10(5), 466-72. [http://www.sciencedirect.com/science/article/pii/S1369526607001173 doi:10.1016/j.pbi.2007.08.008] | ||
| - | + | </div> | |
Latest revision as of 01:22, 27 September 2012
Template:Https://2012.igem.org/User:Tabima
The Basics
Our objective is to generate a genetically-modified bacteria "detect and alert" system, particularly as a defense aid for crop plantations against plant pathogens (1). Bacteria are built (2,4) in a way that they can detect pathogen associated molecules, amplify this initial signal (3), and finally alert the plant (5) by stimulating an early hypersensitive response against infection (6). We have also included a way for the bacteria to control their own population density using a [http://en.wikipedia.org/wiki/Toxin-antitoxin_system Toxin-Antitoxin module] (TA). Using the bacteria Escherichia coli K12, the system will use two differently-modified plasmids. The first one will detect the designated pathogen associeted molecule and weigh such input for intensity and duration (2). If significant, it will also produce a signal molecule to engage the response of the second plasmid (3). Upon exposure to enough concentration of the molecule, the second plasmid will produce a systemic acquired response triggering hormone (salicylic acid) to activate the plant defenses (Loake & Grant, 2007) (5), both attacking the pathogen (6) as well as controlling the bacterial population growth. We strongly believe this biocontrol method may prove to become a powerful tool for farmers and crop workers everywhere. In addition, our constructed "detect and alert" biobricks could be useful to plentiful synthetic biology experiments and future iGEM teams. Finally, want to control against horizontal gene transfer and plasmid curing with using 2 different TA systems.
We're currently implementing our design for the control of Hemileia vastatrix coffee rust by recognizing fungal cell wall chitin ([http://en.wikipedia.org/wiki/PAMP PAMP]), as well as the recognition of the Ralstonia solanacearum 3-hydroxypalmitic acid methyl ester (3-OH-PAME, a [http://en.wikipedia.org/wiki/Quorum_sensing quorum sensing] molecule References (Flavier et al., 1997)) against bacterial wilt. Since the latter is a soil acquired disease, we are also evaluating the possibility of using Pseudomonas fluorescens instead of E. coli as a chassis.
References
- Flavier, A. B., Clough, S. J., Schell, M. A. and Denny, T. P. (1997), Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Molecular Microbiology, 26: 251–259. [http://onlinelibrary.wiley.com/doi/10.1046/j.1365-2958.1997.5661945.x/pdf doi:10.1046/j.1365-2958.1997.5661945.x]
- Loake, G., & Grant, M. (2007). Salicylic acid in plant defence--the players and protagonists. Current opinion in plant biology, 10(5), 466-72. [http://www.sciencedirect.com/science/article/pii/S1369526607001173 doi:10.1016/j.pbi.2007.08.008]
 "
"