Team:University College London/LabBook/Week14
From 2012.igem.org
Sednanalien (Talk | contribs) (→Tuesday 11.09.12) |
YanikaBorg (Talk | contribs) (→Monday 10.09.12) |
||
| Line 9: | Line 9: | ||
| - | '''Method:''' | + | '''Method:''' |
| + | |||
| + | <html><div class="protocol protocol-Generic">Bla bla bla</div><div class="protocolContent"></html>{{:https://2012.igem.org/Team:University_College_London/Protocols/Competency2}}<html></div></html> | ||
| + | |||
| + | |||
| + | https://2012.igem.org/Team:University_College_London/Protocols/Competency2 | ||
== Tuesday 11.09.12 == | == Tuesday 11.09.12 == | ||
Revision as of 00:56, 27 September 2012



Contents |
Monday 10.09.12
Aim: Day 2 of Generating competent cells. The aim is to incubate cells in the presence of CaCl2 to prepare the cell wall, allowing it to become permeable to DNA for transformation.
Method:
https://2012.igem.org/Team:University_College_London/Protocols/Competency2
Tuesday 11.09.12
Aim: Day 3 of Generating competent cells
Method: https://2012.igem.org/Team:University_College_London/Protocols/Competency3

Wednesday 12.09.12
Aim: To carry out a transformation using the 3A ligation products in order to be able to analyse whether ligation was successful.
Method: https://2012.igem.org/Team:University_College_London/Protocols/Transformation1
Thursday 13.09.12
Aim:
Picking of colonies from the ligation transformation plates
Friday 14.09.12
Aim:
To check whether the 3A have carried out the ligation successfully by purifying the O/N culture from the previous day, carrying out an analytical digest and analysing results on a gel.
Methods:
A mini-prep was carried out as per the following protocol:
https://2012.igem.org/Team:University_College_London/Protocols/Miniprep2
This was followed by a 10ul analytical digest as per the following:
https://2012.igem.org/Team:University_College_London/Protocols/EnzDig2
Finally, results of the figest were run on a gel as follows:
https://2012.igem.org/Team:University_College_London/Protocols/Electrophoresis
Results:
The following digram shows results of the analytical digest, with each purified DNA being cut once and twice in order to confirm identity:
| BioBricks | BioBrick Size (bp) | Ligation size (bp) | Plasmid Backbone | Plasmid Expected Size (bp) | Combined Size |
|---|---|---|---|---|---|
| BBa_K729001 & BBA_J23100 | 933 & 35 | 968 | PSB1C3 | 2070 | 3038 |
| BBa_K729001 & BBa_J23119 & BBa_B0034 | 933 & 35 & 12 | 980 | PSB1C3 | 2070 | 3050 |
| BBa_K729002 & BBa_J23119 & BBa_B0034 | 484 & 35 & 12 | 531 | PSB1C3 | 2070 | 2601 |
Conclusion:
The bands on the gel indicate are not as expected. Hence, it is concluded that the ligation was unsuccessful.

Tuesday 11.09.12
Step 1 - Aim:
To generate enough plasmid backbone (PSB1C3) to be used in a 3A ligation. This is followed by a PCR clean-up.
Methods:
https://2012.igem.org/Team:University_College_London/LabBook/Protocols/PCR#Transformation_Protocol_2
Step 2 - Aim:
To check whether PCR is successful. After the PCR reaction, we can detect the presence of the correct products by running a 1% gel electrophoresis.
Methods:
https://2012.igem.org/Team:University_College_London/Protocols/Electrophoresis
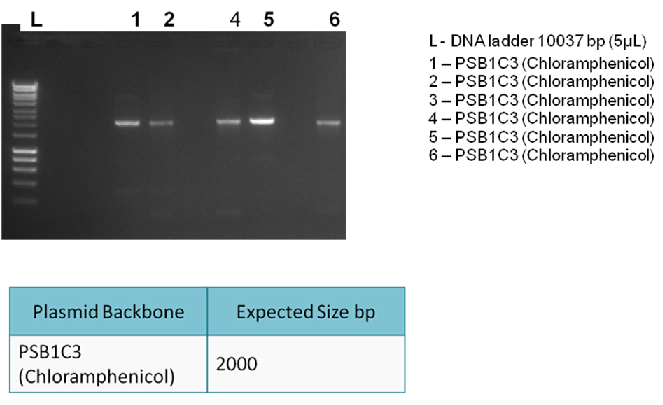
Results:
The following gel results indicate whether PCR was successful.
Conclusion:
Since band sizes were as expected, the PCR was considered to be successful. Hence, the backbone was purified and used in the 3A ligation.

Thursday 13.09.12
Aim:
To generate enough linker (BBa_J23119 + BBa_B0034) to be used in a 3A ligation. This is followed by a PCR clean-up.
Methods: https://2012.igem.org/Team:University_College_London/LabBook/Protocols/PCR#Transformation_Protocol_2
Step 2 - Aim:
To check whether PCR is successful. After the PCR reaction, we can detect the presence of the correct products by running a 1% gel electrophoresis.
Methods:
https://2012.igem.org/Team:University_College_London/Protocols/Electrophoresis
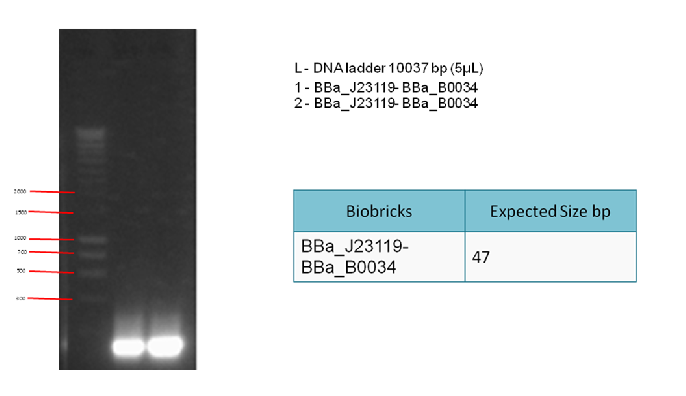
Results:
The following gel results indicate whether PCR was successful.
Conclusion:
Since we obtained the expected bands in the gel, the PCR was considered successful and the purified DNA used in the 3A ligation.

Friday 14.09.13
Aim: To characterise nuclease activity BL21 cells
Method:
To add protocol
Results:
The following table shows readings of the diameters and OD at different time points:
| Date | Time | Colony diameter/mm | Halo diameter/mm | Absorbance at 600 OD |
|---|---|---|---|---|
| Friday | 12.30 | 0 | 0 | 0 |
| Friday | 16.30 | 0 | 0 | 0 |
| Friday | 19.30 | 0 | 0 | 0 |
| Friday | 22.30 | 0 | 0 | 0 |
| Saturday | 01.30 | 0.5 | 1.5 | 0.068 |
| Saturday | 04.30 | 1 | 2 | 0.098 |
| Saturday | 07.30 | 1.5 | 3 | 0.159 |
| Saturday | 10.30 | 1.5 | 3.5 | 0.192 |
| Saturday | 12.30pm | 2 | 4 | 0.203 |
| Saturday | 14.30 | 2 | 4 | 0.209 |
| Saturday | 16.30 | 2.5 | 5 | 0.215 |
The average depth of the colonies was noted to be 0.5 - 1mm
Conclusion:
It can be seen that nuclease works as expected in BL21 cells. However, nuclease activity is less efficient when compared to nuclease activity in WnU cells. This is in line with expectations, since nuclease is found naturally in WnU cells but not in BL21 cells.
 "
"