|
|
| (9 intermediate revisions not shown) |
| Line 4: |
Line 4: |
| | | | |
| | =Week 10= | | =Week 10= |
| | + | |
| | + | All out hard work and preparation in the last weeks came together this week and resulted in great success. Everything ran very smoothly due to our experience and knowledge gained in the lab. It is just as a wise man said 'Everything comes together in the final few weeks'. As a results we were successful in ligating BM and MB with the fluorescent reporters CFP and RFP. This will not only give us two new BioBricks to add to the registry but will also offer us the opportunity to characterise our hybrid promoters in accordance to their function and abilities. |
| | + | |
| | | | |
| | ==Day 1 (10/09/12)== | | ==Day 1 (10/09/12)== |
| | | | |
| - | . As a backup, the running some of the running culture of the Comparator circuit 1 and 2 transformed cells were mini-prepped. After nanodropping, the concentration was found to be:
| + | As backup, the running cultures of the Comparator circuit 1 and 2 transformed cells were mini-prepped. After nanodropping, the concentration were found to be: |
| | | | |
| | C1-1a: 733.4 ng/µL | | C1-1a: 733.4 ng/µL |
| Line 25: |
Line 28: |
| | C2-4a: 811.9 ng/µL | | C2-4a: 811.9 ng/µL |
| | | | |
| - | . Following this an overnight double restriction digest with ''Eco''R1 and ''Pst''1 was set up. These were set up using: 0.2µl BSA, 2µl Buffer H, 0.5µl EcoR1 and 0.5µl Pst1. As to the DNA, 1µg of DNA was added:
| + | Following this an overnight double restriction digest with ''Eco''R1 and ''Pst''1 was set up. These were set up using: 0.2µl BSA, 2µl Buffer H, 0.5µl EcoR1 and 0.5µl Pst1. As to the DNA, 1µg of DNA was added: |
| | | | |
| | C1-1a: 1.36 µL | | C1-1a: 1.36 µL |
| Line 43: |
Line 46: |
| | C2-4a: 1.23 µL | | C2-4a: 1.23 µL |
| | | | |
| - | . Following a low concentration reading on Friday, B-M was miniprepped again. However, this produced no DNA when nanodropped. Therefore, B-M was reinoculated into culture.
| + | Following a low concentration reading on Friday, BM was miniprepped again. However, this produced no DNA when investigated. Therefore, BM was reinoculated into culture. |
| | | | |
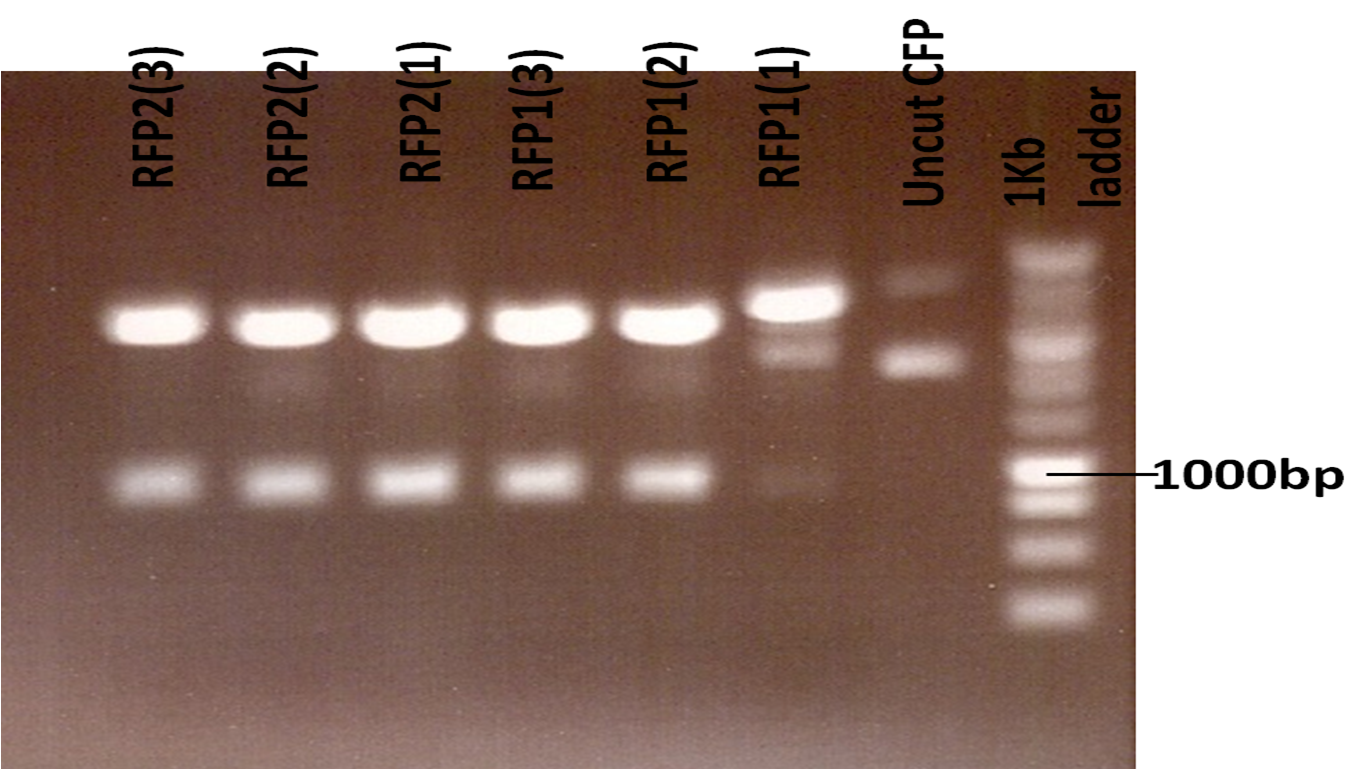
| - | . M-B samples were digested with ''Spe''1 and ''Pst''1 to linearise the backbone to ligate a reporter (CFP, or RFP) into it. The small fragment that was released was discarded during gel purification. At the same time CFP and RFP were digested with ''Pst''1 and ''Xba''1 and then gel purified ready for ligation with M-B. The restriction digests involved 0.5µL of each enzyme, 2µL of Buffer B (''Spe''1 and ''Pst''1)and Buffer H (''Pst''1 and ''Xba''1), 0.2µL of BSA. Again 1µg of DNA was used. The gel used to separate the fragments was 1% w/v. | + | [[File:CFP1.png | thumb | ''Restriction digest of CFP with ''Pst''1 and ''Xba''1'' with the Promega 1kb ladder]] |
| | + | [[File:RFP.png | thumb | ''Restriction digest of RFP with ''Pst''1 and ''Xba''1'' with the Promega 1kb ladder]] |
| | | | |
| - | . Multiple ligations were set up: MB2, MB3,MB5,MB9,MB10 were set up to ligate with both RFP and CFP and Comparator circuit 1 and 2 (all samples) were set up to be ligated in the iGEM backbone pSB1C3. These were all left overnight.
| |
| | | | |
| - | ==Day 2 (11/09/12)==
| + | M-B samples were digested with SpeI and PstI to linearise the backbone in order to ligate a reporter (CFP, or RFP) into it. The desired DNA fragment was [https://2012.igem.org/Team:NRP-UEA-Norwich/Protocol#Purification_by_Centrifugation gel purified]. At the same time, CFP and RFP were cut with PstI and XbaI and then gel purified to be ready for ligation to MB. The restriction digests involved 0.5µL of each enzyme, 2µL of Buffer B (SpeI and PstI)and Buffer H (PstI and XbaI) and 0.2µL of BSA. Again 1µg of DNA was used. The agarose gel in which the fragments were separated was of 1% w/v. |
| | | | |
| - | . The ligations (MB with CFP/RFP and Comparator Circuit 1 and 2 with pSB1C3) from the night before were run on a 1% w/v gel for an hour and the fragments were cut out. These gel slices were purified. The purified DNA, was then transformed into Bioline Alpha Select Gold Standard Competence cells. The plates were left overnight in an incubator at 37 degrees Celsius.
| + | The DNA samples used were MB2, MB3,MB5,MB9,MB10. Following a successful ligation, the backbones containing the hybrid promoters were ready to be ligated either RFP or CFP. I preparation of constructing our Comparator circuit 1 and 2, the synthetic, complementary sequences were set up to be ligated in the iGEM backbone pSB1C3. All reactions were left overnight. |
| | | | |
| - | . Samples of B-M were miniprepped and nanodropped. The concentrations were found to all be lower than 45ng/L. Furthermore, the 260/230 readings were all negative.
| |
| | | | |
| - | . Digested:
| + | ==Day 2 (11/09/12)== |
| | | | |
| - | '''working mathmatical modelling'''
| + | The ligations (MB with CFP/RFP and Comparator Circuit 1 and 2 with pSB1C3) from the night before were run on a 1% w/v gel for an hour and the fragments cut out. The gel slices were then purified. The obtained DNA samples were then transformed into Bioline Alpha Select Gold Standard Competence cells. The plates that were being produced in the process were incubated at 37 degrees Celsius over night. |
| - | [[File:Comparator_circuit_equasion_iGEM_1_12.09.15.jpg | 200px | right]]
| + | |
| | | | |
| - | E = the expression of one of the fluorescent protiens (RFP) when there is transcription of the CFP RNA at any particular level as a proportion of the expression of RFP at the same transcription rate when none of the CFP RNA is present within the cell. So if the promoter (PYEAR) attached to the rfp and construct 1 was expressing at a constant rate with promoter 2 entirely switched off then promoter 2 started transcribing and the amount of rfp in the cell halved then the value of EA would be 0.5 at that transcription rate of CFP (0-1)
| + | Samples of BM were miniprepped and nanodropped. The concentrations were found to all be lower than 45ng/L. Furthermore, the 260/230 readings were negative. |
| | | | |
| - |
| |
| - | L = the length of the DNA strand that is transcribed (Leader and protein coding region) .
| |
| - |
| |
| - |
| |
| - | C = the rate of transcription
| |
| - |
| |
| - |
| |
| - | L/C = the period of time taken for transcription to take place, the time in which translation can be initiated but it is unlikely that the two leaders will bind to one another
| |
| - |
| |
| - |
| |
| - | A = the rate of transcription of promoter 1 (the PYEAR) as a proportion of it’s maximum possible transcription rate (0-1)
| |
| - |
| |
| - |
| |
| - | A*(L/C) = the number of RFP RNAs that can be translated independently of the presence of other RNA at any one time and so is proportional to translation (and expression) from DNA ascociated RNA (RNA that is still being transcribed). This occurs both when the CFP RNA is and isn’t present so has to be on both the top and bottom of the equation.
| |
| - |
| |
| - |
| |
| - | H = Half life of the RNA after transcription
| |
| - |
| |
| - |
| |
| - | L/C + H = the full time for which the RNA would be translated assuming no interactions between leaders for instance when only one of the promoters is inducing transcription.
| |
| - |
| |
| - |
| |
| - | B = the transcription of the second promoter within the cell as a proportion of the maximum possible transcription of that promoter (0-1)
| |
| - | Because there can not be negative expression of A (only positive expression of B to represent negative expression of A) the translation of the RFP RNA(A) = A - AB .
| |
| - | It also has to be remembered that there will never be full interaction between the two RNA leaders, particularly at low concentrations if only because the two strands never come in to proximity or because of cellular processes; consequently the function B/(D+B) must be used giving the formula; translation of the RFP RNA(A) = A – A (B/(D+B))
| |
| - |
| |
| - |
| |
| - | D is a constant of the biological system whose derivation is so complex that it can only really be calculated through observation but can be modelled at various levels.
| |
| - |
| |
| - |
| |
| - | So to bring it all together; the top half of the equation indicates the degree of translation of the RNA transcribed by the first promoter under any particular transcription rate of the two promoters in arbitrary units. To make this into a meaningful output it is divided by the maximum translation rate at that rate of transcription to equal EA ; this indicates the degree of attenuation of one RNA from the other.
| |
| - |
| |
| - |
| |
| - | To get the degree of translation of the other RNA (EB) just swap A for B throughout the equation.
| |
| | | | |
| | ==Day 3 (12/09/12)== | | ==Day 3 (12/09/12)== |
| | | | |
| - | . A B-M overnight culture was miniprepped by Russell, and nanodropped. The concentration was found to be: 199 ng/µL
| + | A BM overnight culture was miniprepped by Russell and nanodropped. The concentration was found to be 199 ng/µL. The DNA was then digested with SpeI and PstI. Again 1µg of DNA, 0.5µL of each of SpeI and PstI, 0.2µL BSA and 2µL Buffer B were added to each digest. Following the incubation period, the sample were run on a 1% w/v agarose gel for an hour and the large fragment purified. I addition multiple overnight ligations were set up for BM1 and BM4 with RFP and CFP. It was made sure that the samples remained in the dark. |
| | | | |
| - | . This was then digested with ''Spe''1 and ''Pst''1. Again 1µg of DNA, 0.5µL of each of ''Spe''1 and ''Pst''1, 0.2µL BSA and 2µL Buffer B. | + | All the plates, which showed growth from the previous days experiments, were inoculated into 5ml LB cultures containing the relevant antibiotics. Furthermore, as back up, Comparator circuit 1 and 2 were transformed again and plated. These plates were grown overnight at 37 degrees Celsius. |
| | | | |
| - | . This was then run on a 1% w/v agarose gel for an hour and the large fragment purified.
| |
| | | | |
| - | . Multiple overnight ligations were set up for BM1 and BM4 with RFP and CFP. These were left in the dark.
| + | ==Day 4 (13/09/12)== |
| | | | |
| - | . All the plates which showed growth were inoculated into 5ml LB cultures containing the relevant antibiotics. | + | Inoculations were prepared of BM and MB2 to CFP and GFP. These were inoculated into several LB cultures with chloramphenicol added. We also transformed BM and incubated at 37 degrees Celsius over night. |
| | | | |
| - | . As back up, Comparator circuit 1 and 2 was transformed again and plated. These plates were grown overnight at 37 degrees Celsius.
| |
| | | | |
| | + | ==Day 5 (14/09/12)== |
| | | | |
| - | | + | Samples of MB2 attached to RFP and CFP were transformed and plated. These plates were incubated over the weekend. We then re-inoculated samples of BM and MB2 ligated to CFP and RFP. |
| - | ==Day 4 (13/09/12)==
| + | |
| - | | + | |
| - | ==Day 5 (14/09/12)==
| + | |
 "
"