Team:Amsterdam/achievements/stochastic model
From 2012.igem.org
(Difference between revisions)
(→Introduction) |
|||
| Line 51: | Line 51: | ||
==ODE Model definition== | ==ODE Model definition== | ||
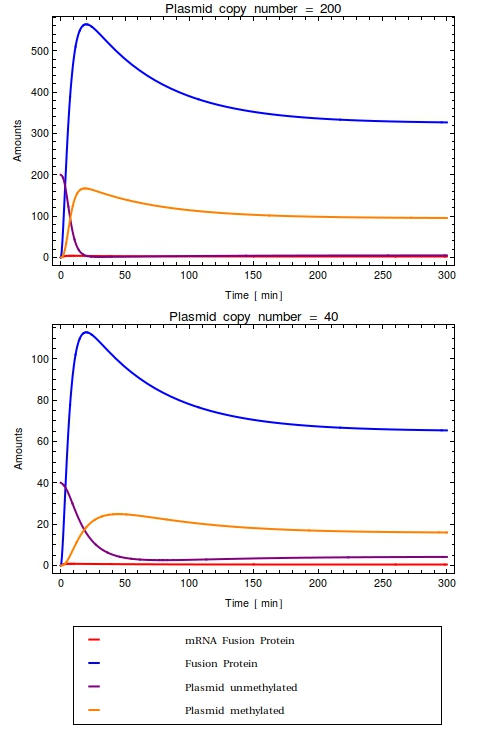
| + | [[File:Combinedode.png|thumb|right|300px|Time trajectories of ODE-model in which the construct has been inserted in a high copy number plasmid (top, $Ca = 200$) and a low copy number plasmid (bottom, $Ca = 40$). All initial species values are set to $0$, except for the intial value of unmethylated plasmids which is equal to $Ca$. With these parameters, the dynamic range of the system is completely taken up by the background noise. No qualitative difference is observed between the high and low copy number cases]] | ||
| + | |||
FP-mRNA results only from leaky expression from the operons $O$ with leakiness rate $k_{\text{sMFP}}$ and is degraded with rate $\lambda_{\text{MFP}}$: | FP-mRNA results only from leaky expression from the operons $O$ with leakiness rate $k_{\text{sMFP}}$ and is degraded with rate $\lambda_{\text{MFP}}$: | ||
$$ | $$ | ||
| Line 121: | Line 123: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | |||
Looking at the plots for both the high and the low copy number plasmids, no qualitative difference is observable. All species in the system simply reach a steady state defined as the fraction between their production and degradation rates. | Looking at the plots for both the high and the low copy number plasmids, no qualitative difference is observable. All species in the system simply reach a steady state defined as the fraction between their production and degradation rates. | ||
Revision as of 20:27, 26 September 2012
 "
"