Team:Paris-Saclay/Project/Notebook/Week 8
From 2012.igem.org
(Difference between revisions)
YohannPetiot (Talk | contribs) |
YohannPetiot (Talk | contribs) |
||
| Line 23: | Line 23: | ||
</head> | </head> | ||
<body> | <body> | ||
| - | <div id="single- | + | <div id="tiles"> |
| - | <div | + | <div id="single-tile3" class="red live-tile" data-mode="flip" data-delay="4000"> |
| - | + | <div> | |
| - | + | <a href="https://2012.igem.org/Team:Paris-Saclay/Project/Abstract"> | |
| - | + | <div class="child-tile"><p class="child-tile">GEMOTE Project</p></div> | |
| + | <img src="http://www.igem-paris-saclay.u-psud.fr/wordpress/wp-content/uploads/2012/07/Aud.jpg" alt="" /> | ||
| + | </a> | ||
| + | </div> | ||
<div> | <div> | ||
| - | <img src="http://www.ecole-adn.fr/uploads/2011/09/Probiotics-300x300.jpg" alt="" /> | + | <a href="https://2012.igem.org/Team:Paris-Saclay/Project/Abstract"> |
| - | + | <img src="http://www.ecole-adn.fr/uploads/2011/09/Probiotics-300x300.jpg" alt="" /> | |
| + | <div class="child-tile"><p class="child-tile">GEMOTE Project</p></div> | ||
| + | </a> | ||
</div> | </div> | ||
| - | <script type="text/javascript"> | + | </div> |
| + | <script type="text/javascript"> | ||
// apply regular slide universally unless .exclude class is applied | // apply regular slide universally unless .exclude class is applied | ||
// NOTE: The default options for each liveTile are being pulled from the 'data-' attributes | // NOTE: The default options for each liveTile are being pulled from the 'data-' attributes | ||
$(".live-tile, .flip-list").not(".exclude").liveTile(); | $(".live-tile, .flip-list").not(".exclude").liveTile(); | ||
| - | </script> | + | </script> |
| - | </div> | + | </div> |
</body> | </body> | ||
</html> | </html> | ||
Revision as of 14:46, 26 September 2012
23rd July
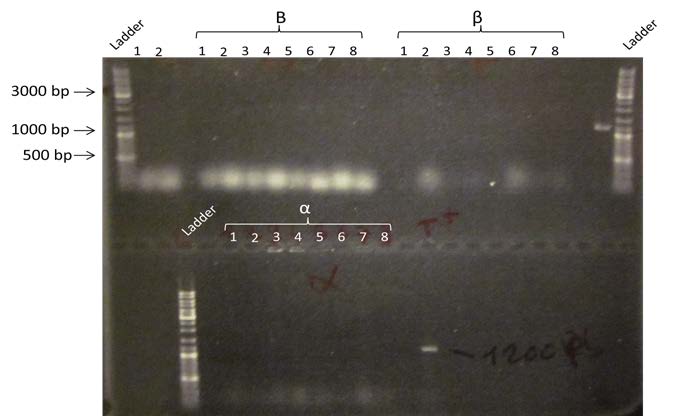
| Electrophoresis of the post Gibson assembly PCR on a 1% Agarose gel. We are expecting a band at 4800bp for B, and a band at 1200bp for α and β. | |
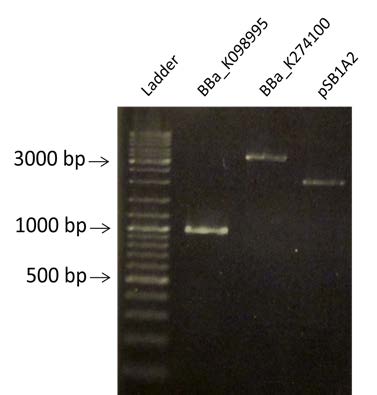
| Checking of the purity of BBa_K098995, BBa_K274100 and pSB1A2, all already digested by DPNI |
24th July
- Purification by column of BBa_K098995, BBa_K274100 and pSB1A2.
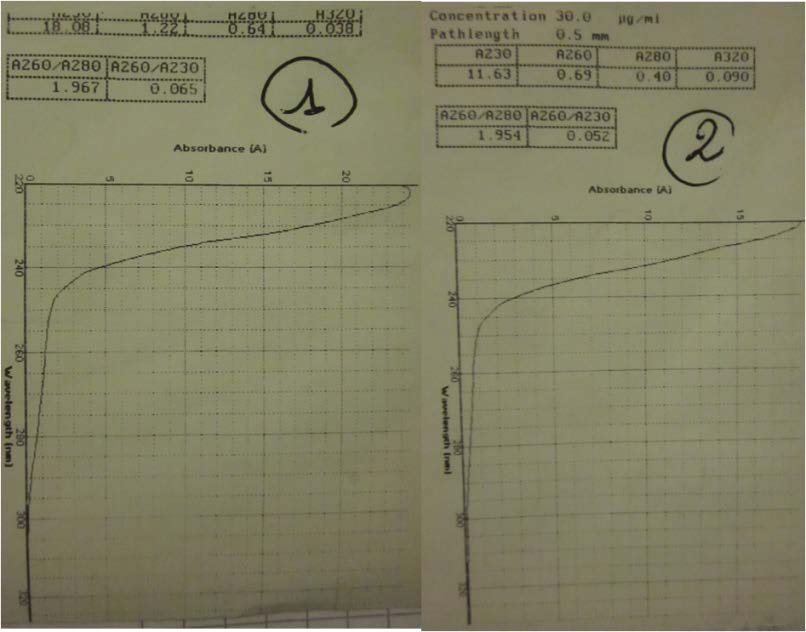
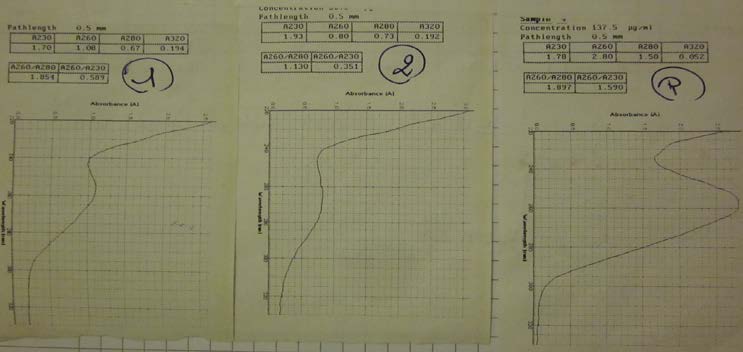
- Estimation of the concentration of BBa_K098995, BBa_K274100 and pSB1A2 by Nanovue.
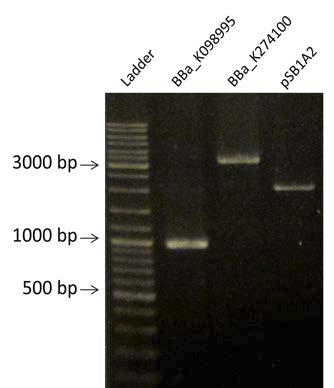
| Visualization of the purification of BBa_K098995, BBa_K274100 and pSB1A2 by electrophoresis on a 1% Agarose gel. We are expecting a band at 935 bp for BBa_K098995, 3385 pb for BBa_K274100 and 2079 bp for pSB1A2. |
25th July
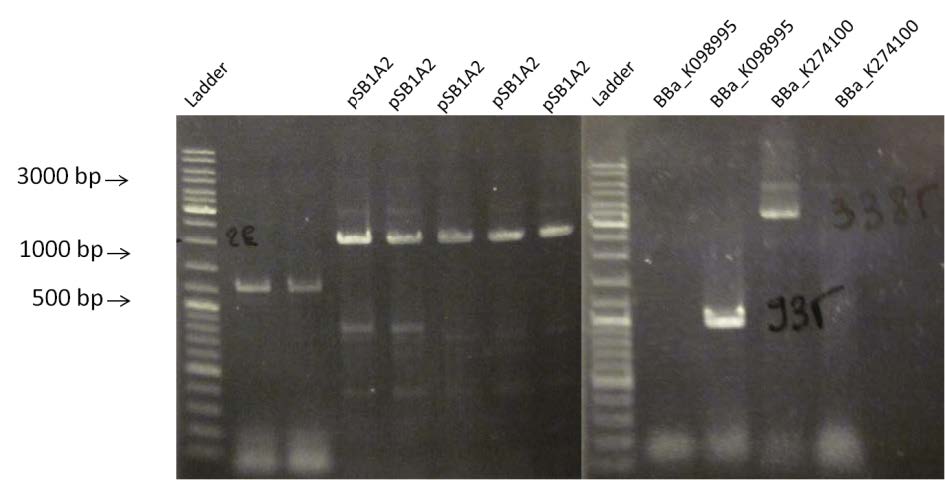
- Amplification by PCR of BBa_K098995, BBa_K274100 and pSB1A2. Visualization by electrophoresis on a 1% Agarose gel. We are expecting a band at 935 bp for BBa_K098995, 3385 pb for BBa_K274100 and 2079 bp for pSB1A2.
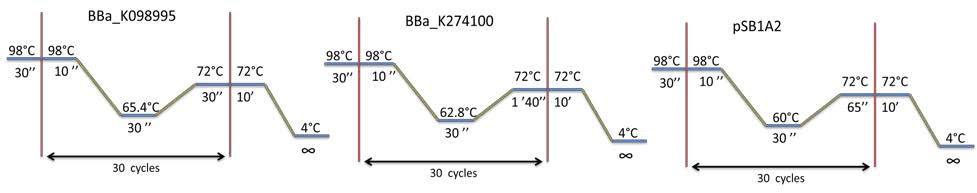
PCR program used:
- Treatment by DPNI on BBa_K098995, BBa_K274100 and pSB1A2 in order to eliminate the plasmid matrix.
26th July
- Purification of BBa_K098995, BBa_K274100 and pSB1A2 with the gel extraction PCR clean up kit to prepare a Gibson assembly.
- Nanovue to measure the concentration of our 3 samples: BBa_K098995, BBa_K274100 and pSB1A2.
27th July
- Gibson assembly for the B construction (BBa_K115017 + 880bp end of BBa_K098995 + BBa_K274100 + BBa_J61048 + pSB1A2). A control is done with just the plasmid treated by DPNI.
- Transformation of competent cell DH5α with the B construction. Spreading out on LB+Agar+Ampicilline, and the Petri dishes are placed at 37°C.
 "
"


Follow us !