Team:Peking/Project/Luminesensor/Design
From 2012.igem.org
m |
|||
| Line 141: | Line 141: | ||
<h3 id="title5">Reference</h3> | <h3 id="title5">Reference</h3> | ||
<p></p> | <p></p> | ||
| - | <ul><li id="ref1"> | + | <ul class="refer"><li id="ref1"> |
1. Shimizu-Sato, S., Huq, E., Tepperman, J.M., & Quail, P.H.(2002). A light-switchable gene promoter system. <i>Nat. Biotechnol.</i> 20: 1041: 1044 | 1. Shimizu-Sato, S., Huq, E., Tepperman, J.M., & Quail, P.H.(2002). A light-switchable gene promoter system. <i>Nat. Biotechnol.</i> 20: 1041: 1044 | ||
</li><li id = "ref2"> | </li><li id = "ref2"> | ||
Revision as of 10:30, 26 September 2012
Summary
This summer, our team wished to create a novel optogenetic tool that could really make a big difference in the field of synthetic biology. But what should the difference be? In order to answer this question, we should perform a brief retrospect to meditate upon what accomplishment has already been achieved and what problems remained to be solved in optogenetic research.
Design: A Retrospect
As we discussed before, virtually all of current light-sensitive optogenetic modules have been designed following two similar general principles –- attaching a physiologically functional domain to a photoreceptor domain or rewiring physiological pathways to the downstream of the signaling pathways of photoreceptor proteins in order to use light to trigger physiological response. These two principles, whose concepts easy to understand, actually bear in their kernel the spirit of modular design which has been so strongly proposed in synthetic biology, with the former principle particularly versatile and easy to manipulate.
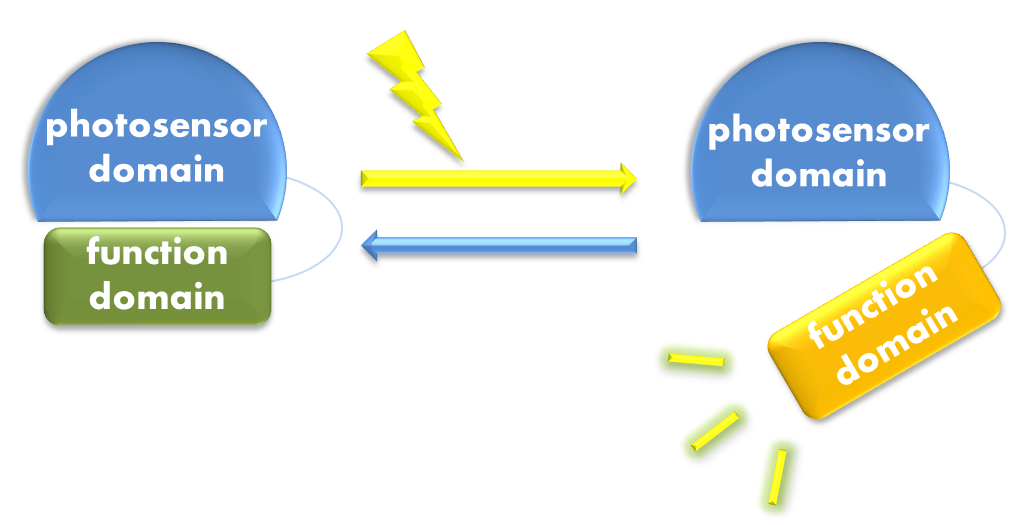
Figure 1. Illustration of the first general principle of optogenetic design. Designs of fusion proteins in optogenetic research have followed a general principle of attaching a physiological functional domain to a photosensor domain. When light excites the photosensor domain to trigger, say, confromational change, the change will be transmitted to the physiological domain to induce physiological effect.
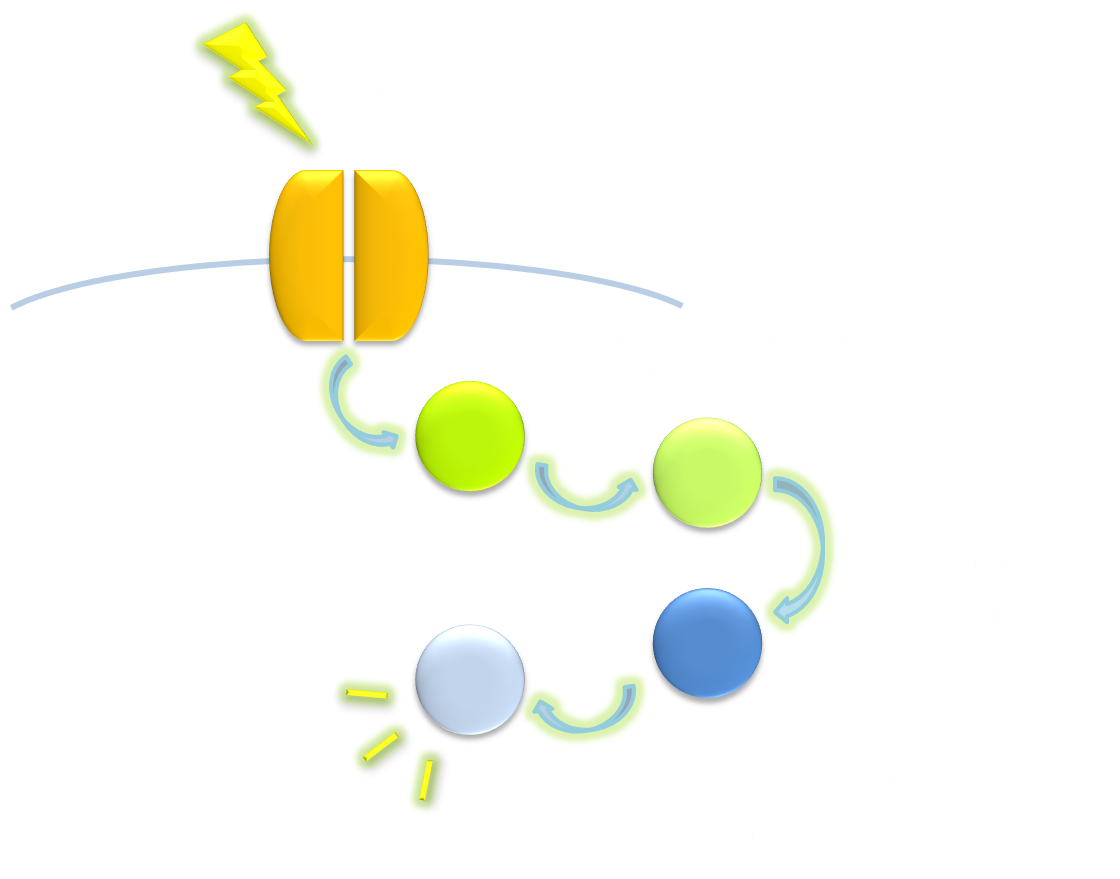
Figure 2. Illustration of the second general principle of optogenetic design. There is another principle for optogenetic design, namely to rewire a physiological signaling pathway to the downstream of the signaling pathway of a photosensor. Thus the physiological effect of the physiological pathway may occur as a ramification of the photosensor signaling cascade.
Thus we reasoned that the first principle is an appropriate paradigm to follow. However, in order to make our novel optogenetic tool an unprecedented success, the functional domain, photoreceptor domain and the connection between them must be carefully chosen and sophistically designed to solve several important problems that have become gradually manifest along with the development of optogenetic research, for they may impede the future application of our optogentic tool.
First of all, in order to be widely applicable for biotechnological use, a photosensitive module must be truly ‘sensitive’. Some of the existing photosensitive modules work best around light intensity of 10W/m2, which is about the illuminance inside a room on a sunny day; some even require laser beam to activate, whose light intensity can hardly be achieved in natural environment. Apparently, the dependency on high light intensity is severely defective, because high-intensity light could be detrimental to cellular function or can even cause cell death. Moreover, this defect limits the application of optogenetics in future possible scenarios where photosensitive modules may be required to respond to soft light that is cell-and tissue-friendly, or light emitted by biological organisms, or, even more stringent, light emitted by bacteria, which is by no means brighter than the dim light of the moon.
Secondly, the photosensitive modules must be widely compatible. Most of current photosensitive modules have utilized light-sensitive domains of photoreceptors of eukaryotic origin, which often fail to work in prokaryotic organisms. Yet comparing to eukaryotic organisms, prokaryotic cells require significantly less stringent culturing conditions and are considerably less susceptible to environmental pollutions. What’s more, some of current photosensitive modules utilize photoreceptor domains such as phytochromes where exogenously supplied specific chormophores are necessary for its normal function. These two obvious defects substantially block the way to applying optogenetics to synthetic biology where in-field application is very much considered.
Thirdly, the design of a successful photosensitive module should be instructive to future photosensitive module engineering, which is particularly important for synthetic biology. In another word, its design should be modular enough to make possible the reshuffling of functional domain to construct modules with novel physiological functions. With no much deduction, one can easily postulate that this would require equal amount of modularity in the original photosensor domain and physiologically functional domain based on which the design was constructed.
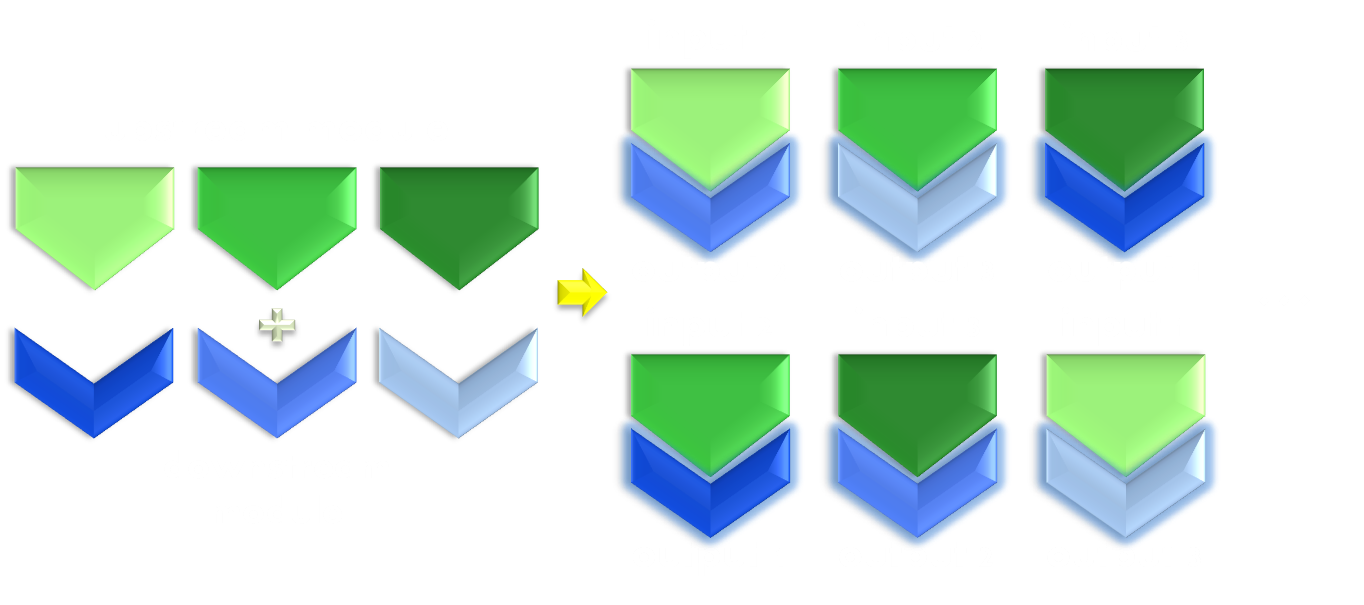
Figure 3. Illustration of the importance of modularity. Modularity is important in both evolution and synthetic biology. As is shown in the figure, even if only equipped with limited amount of modular parts, we can interchange the relationship between them to create many more new physiological function. Thus designs possessing modular components would be much more enabling than a non-modular one.
Now, with the little retrospect above, we have determined the ‘differences’ we would like to make: sensitivity, compatibility and modularity. But how to create a photosensitive optogenetic tool that is ultra-sensitive, widely compatible and highly modular all at the same time? To address this problem, we need to start from choosing a promising light-sensitive photoreceptor domain—the core of our design.
Design: Choosing the Photoreceptor
Domain
From the discussion above, we have already formed a few criteria that the photoreceptor domain must satisfy. It should have been proved to be highly sensitive, should incorporate no chromophore that cannot be synthesized by bacteria, and should be modular in structure and function. Based on these standards we can go through the photoreceptor proteins previously used in optogenetic research, examine them one by one, and rule out inappropriate ones one by one to finally reach the most proper light-sensitive photoreceptor domain for our design.
The light-sensitive domains of most of existing optogenetic modules can be categorized into three major groups: rhodopsins, phytochromes, and phototropins, each with distinct characters and utilities.
Rhodopsins are membrane anchored proteins that can be devided into two subtypes. Both of them detect light through a retinal chromophore. Type 1 rhodopsins, with bacteria rhodopsin as a prototypical representative, directly drive ionic currents through a cycle of light-induced conformational change. Type 2 rhodopsins, with bovine rod rhodopsin as a representative, regulate ionic currents through a downstream signaling pathway triggered by light-induced conformational change in the molecule. The type 1 rhodopsins proved very useful in neuron science. For example, by expressing Channelrhodopsin-2 (ChR2), a light-sensitive ion-channel from green algae which will become permeable to cations after being triggered by blue light, under the control of a tissue specific promoter, scientists have achieved precise and timed light-dependent activation of specific neuronal cells. Type 2 rhodopsons can also achieve light-dependent spatial-temporal control of gene expression through synthetic rewiring of calmodulin signaling pathway which will activate nuclear transcription to the downstream of signaling pathway of rhodopsins such as melanopsin, which will, upon light activation, trigger calcium influx. However, rhodopsins may not be suitable for our project, partly due to the extraordinary integrity of its structure and its membrane-anchored nature, which makes it difficult to fit into the framework of the first principle we have mentioned before, and partly due to the requirement of exogenous supply of the chromophore—retinal, which is not involved in common bacteria metabolism pathways. These two unique features clearly contradict with our goal—‘modularity’ and ‘compatibility’, and thus ruled rhodopsin out of our consideration.

Figure 4. Illustration of function mechanism of type 1 rhodopsin. Rhodopsin has an invariant retinal chromophore. When excited by light, the retinal molecule will undergo conformational change from a cis-conformation to a trans-conformation, and causes subsequent conformational change in rhodopsin molecule and then triggers downstream effect. In type 1 rhodopsin, conformational change in rhodopsin molecule will induce ion influx.
The plant phytochrome B (PhyB) is an outstanding representative of phtochrome photosensors. PhyB photoconverts between two forms: when PhyB in its inactive form (Pr) absorbs red light, it converts into its active form (Pfr), and reconverts back into Pr form slowly, when left in the dark, or rapidly, when exposed to far red light. In its physiologically active form Pr, PhyB is able to interact with its nuclear signaling partener Phytochrome Interacting Factor 3 (PIF3) to form a dimer. By fusing DNA binding domain and transcription activating domain of GAL4 to PhyB and PIF3, respectively, scientists have been able to create a light based yeast-two-hybrid system. As a variation of the theme, scientists have managed to utilize this light-dependent protein-protein interaction to recruit cell signaling molecules to the plasma membrane of mammalian cells to trigger responses such as pseudopod formation and cell polarization. Although radiating with such promise, the phytochrome still cannot be included in our system, simply because it requires, again, the exogenous supply of chromophore (in the PhyB’s case, phycocyanobilin (PCB) ). Although solutions such as incorporating into cells metabolic pathways which will enable them to synthesize these chromophores have been proposed, these proposals will greatly complicate the process of design and application of the light-sensitive system. Although Phytochormes have been excluded from our plan, its mechanism greatly enlightened us. Those previous designs that have utilized the heterodimerization of PhyB and PIF3 have grabbed the essence of a common theme in the functioning of cellular processes—the protein-protein interaction, and have been exemplary in demonstrating a modular and versatile design. These particular characters are precisely the things we are looking for. When we are going further to seek our ideal photosensor domain, we shall keep this example in mind.
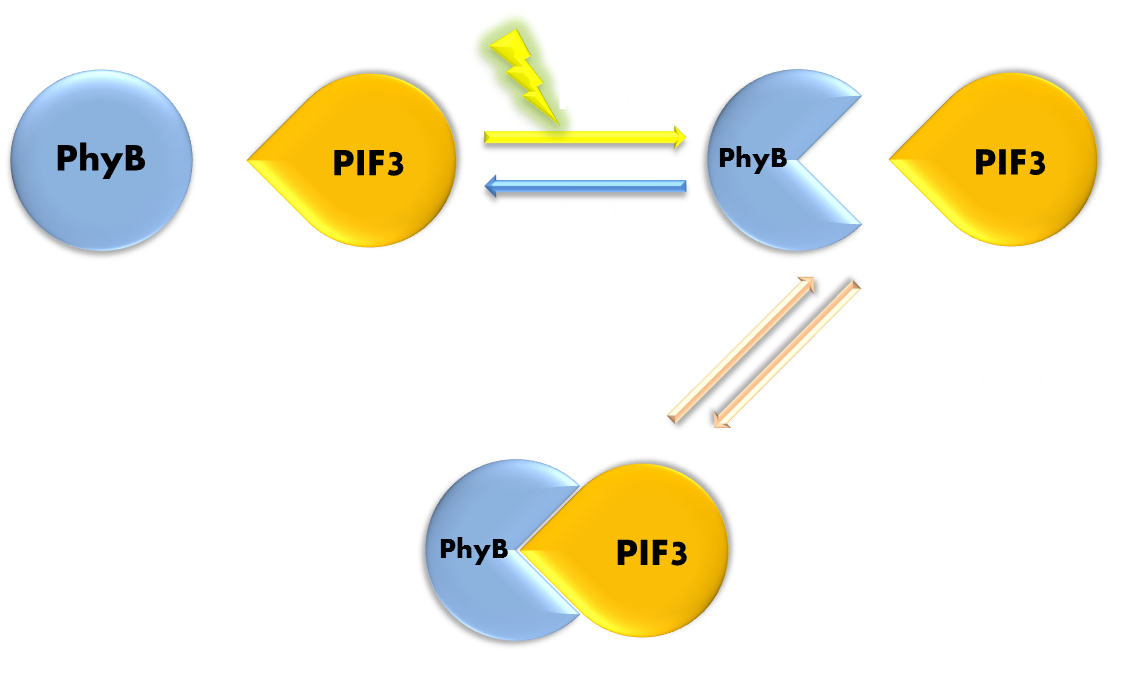
Figure 5. Illustration of function mechanism of phytochrome PhyB-PIF3 system. PhyB-PIF3 system is a outstanding representative of phytochorme photosensors. PhyB utilizes a phycocyanobilin (PCB) molecule as chromophore. When exposed to red light, PhyB will convert from its Pr (inactive) form to its Pfr (active) form. Pfr can bind to PIF3 and form a heterodimer. When exposed to far-red light or kept in the dark, PhyB will convert back to its Pr form, and the dimer will disassociate.
Now we have come to phtotropins. We gladly found that among all these three groups of commonly used light-sensitive domains, phototropins possesses the most distinct modular structure. Phototropins have a structurally conserved light sensor domain, termed LOV (light, oxygen and voltage) domain, which is easily discernable and often precedes, within a single reading frame, a sequence of an enzymatically functional domain, connected by the sequence of a linker domain. Despite its structural modularity, the LOV light sensitive domain also seems to possess an outstanding functional modularity, for it was found to combine with various functional domains to perform various cellular functions. Several classes of LOV domain–effector domain combinations have hitherto been described, such as LOV-HKs combination, performing histidine kinase function, LOV GGDEF–EAL combination, predicted to regulate the synthesis and hydrolysis of cyclic di-GMP, and LOV_HTH combination, performing light-induced DNA binding activity. Such versatility already existing in nature strongly suggested that a newly designed artificial fusion protein that fuses the LOV photosensitive domain to a physiologically functional domain to create a non-pre-existing combination would probably work. LOV photosensor domain seems to bear much more virtue than just its modularity. LOV domains have a non-covalently bound flavin (FMN or FAD) chromophore that is absolutely essential for its function. Unlike those choromophres of rhodopsins and phytochormes, flavin is an essential chemical compound deeply involved in the respiratory chain in all forms of life, including bacteria. In another word, in order to let this LOV domain function in bacteria, we only need to incorporate the gene into the cell, and the cell will automatically supply the protein with the chromophore.
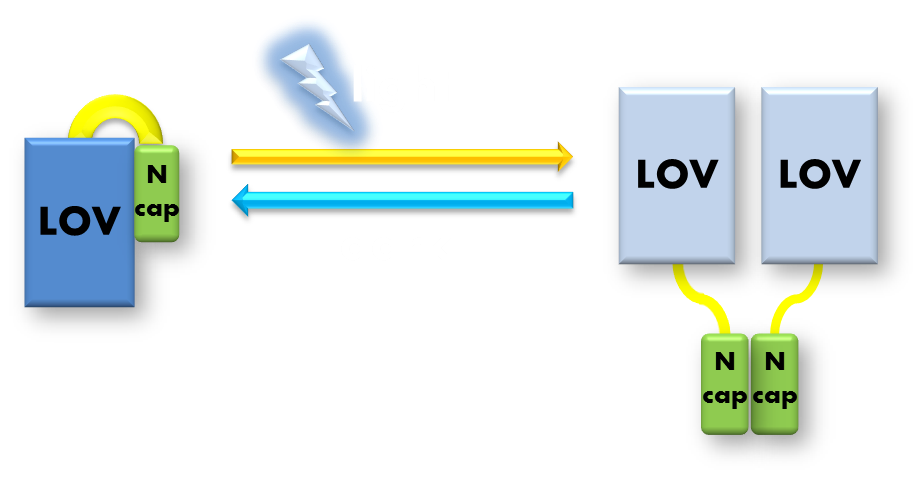
Figure 6. Illustration of function mechanism of phototropin VVD. VVD belongs to phototropin photosensor family. VVD posseses a clearly distinguishable photo-sensitive LOV domain and utilizes a FAD (flavin adenine dinucleotide) molecule as its chromophore. When excited by blue light, the N-terminal cap of VVD protein will disassociate from the LOV domain and serves as a dimerization surface for subsequent homo-dimerization. When the dimerized VVD proteins are no longer exposed to blue light, they will gradually decay back to its dark state.
So far our good phototropin has satisfied two of our requirements: modularity and compatibility. So how about its sensitivity? To date, scientists have, for example, attached Rac1 protein to LOV2 domain from phototropin1 originated from Avena sativa to achieve photoactivatable cell motility, or shuffled the histidine kinase domain of FixL protein, which belongs to a two-component system, to the downstream of the LOV domain of B. subtilis YtvA protein to create a light-regulated histidine kinase, and so on. Among these designs, a particular one that utilizes the photosensor domain of a Vivid (VVD) protein, which originated from Neurospora. Crassa, camptured our attention. The VVD-GAL fusion protein thus designed achieved a rather high light-sensitivity—about 0.04W/m2. This is definitely thrilling news. Moreover, VVD was shown to form a rapidly exchanging homodimer upon blue-light activation, which means, like the Phy-PIF system, the general theme of protein-protein interaction might also be applied to this particular protein. Adding more promise to this VVD protein is its size—the smallest LOV domain containing protein known. This feature makes it easy to engineer and more likely to be stably expressed in bacteria systems.
Based on the discussion above, we have finally chosen the smallest LOV protein (or phototropin) VVD protein’s photosensor domain as our photoreceptor domain. Due to its small size, structural and functional modularity, bacteria compatibility, previous experience of being highly sensitive and its photoswitching mechanism, we reasoned that VVD photosensor domain would be an enabling tool in our coming design of a novel optogenetic module.
Our next step is to choose a physiologically functional domain for our new optogenetic module.
Design: Choosing the Physiologically
Functional Domain
Now we have to choose a physiologically functional domain for our optogenetic module.
To do this, we need to decide in first place what physiological function we would like our module to achieve. Considering the limited amount of time we have, we need the function of our light-sensitive module easy to manipulate and characterize; and to make our light-sensitive module widely applicable in biotechnological field, we need its function to be modular. So we choose the most conventional and fundamental way of controlling cellular behavior as our module’s function: tuning gene expression through transcriptional control.
In bacteria system, transcriptional control of gene expression is often achieved trough the association and disassociation of transcriptional factors (TF) with target sequence on or around promoter site of a particular gene. Among these bacteria Transcriptional Factors, transcription inhibitors take up the major proportion and, coincidentally, many of the inhibitors require dimerization for its inhibitory function. What’s more, these transcriptional factors that require dimerization to function usually consist a DNA binding domain. Now, based on these facts, we can envisage a rough blueprint of our design: the DNA binding domain of a transcription inhibitor shall be fused to the downstream of VVD photosensor domain, and upon blue-light activation, VVD domain will dimerize, helping the DNA binding domain dimerize at the same time to enable its DNA binding activity and inhibition of transcription initiation.
But what transcription factor should we use exactly? In order to work well with our VVD photosensor domain, the DNA binding domain of the transcriptional factor must be easily discernable in the whole structure and had better been proved to function independent of the rest part of the protein. Of course, the DNA binding activity of the transcriptional factor we choose must be specific and rigorously require dimerization.
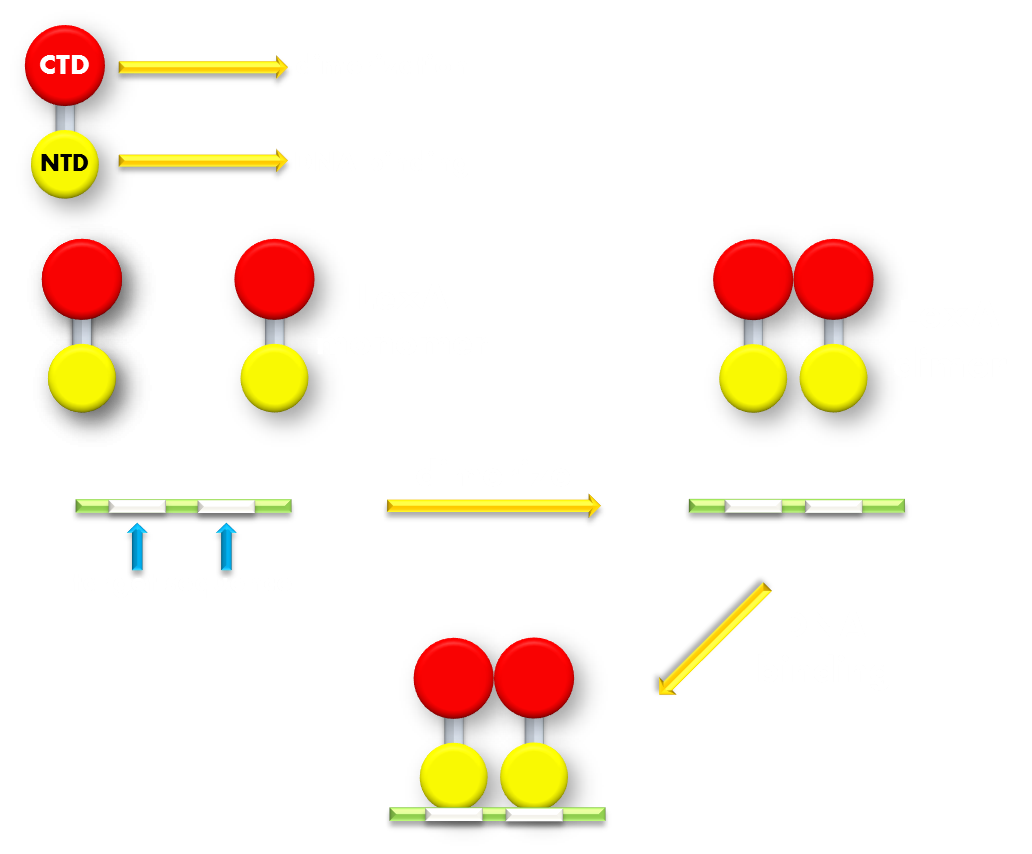
Figure 7.Illustration of function mechanism of LexA transcription repressor. LexA is the general repressor of the genes in bacteria SOS system. It is composed of a C-terminal domain (CTD), which is responsible for dimerization, and a N-terminal domain (NTD), which is responsible for DNA binding. SOS system genes possess a (some possess several) symmetrical sequence (CTGT(N)8ACAG) in their promoters, which is called the SOS box. LexA proteins will form a dimer in bacteria cell and the NTDs will recognize and bind to the SOS box in SOS promoters and thus inhibit transcription initiation.
Based on these criteria, a transcription repressor,LexA, came into our sight. LexA is a transcription repressor of all the genes in the SOS system in E.coli, and its crystal structure has been resolved at high-resolution. LexA protein consists of a N-terminal DNA binding domain and a C-terminal dimerization domain with a short hydrophilic linker linking the two separate domains, making the structure impressively modular. Under normal physiological conditions, two LexA proteins will form a homodimer through the dimerization domain interface and bind to the SOS box target gene sequence in promoters of genes in SOS system and create significant steric hindrance for transcription polymerase binding and thus inhibit transcription initiation.
From these facts, we can conclude that the LexA repressor is the suitable physiological functional domain for our light-sensitive module, as it matches all of our criteria. First of all, a high-resolution crystal structure clearly shows its structural modularity. Beside the apparent structural modularity, various researches have utilized LexA to create versatile gene expression regulating modules that have convincingly demonstrated its functional modularity, further adding to its promise. (For example, a LexA-based genetic system has been devised for monitoring and analyzing protein heterodimerization in Escherichia coli.) Moreover, the DNA binding domain of LexA rigorously requires dimerization to bind efficiently to its targeting site (SOS box), and DNA binding domain itself has no dimerization ability. Last but not least, LexA is bacteria endogenous protein, which means it is likely to be efficiently expressed and function normally in bacteria systems.
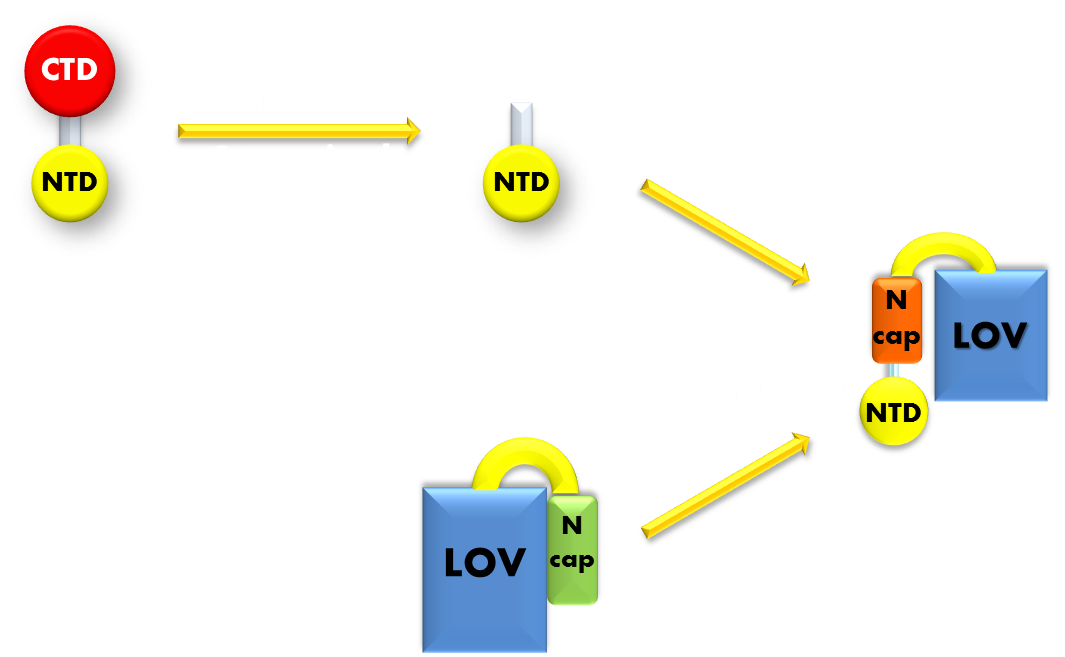
Figure 8. Procedure of building up our luminesensor. We take the N-terminal domain of LexA protein, which is responsible for DNA binding, and fused it to the N-terminal of VVD photosensor protein.Thus our luminesensor is constructed.
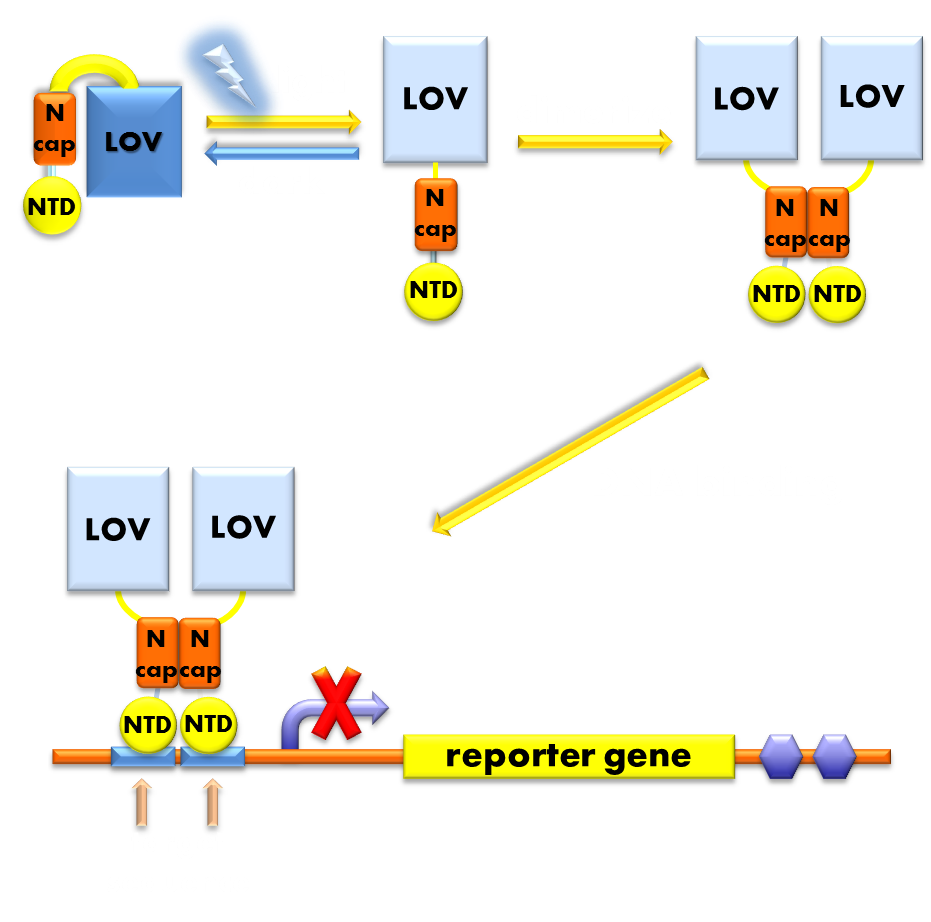
Figure 9. Illustration of the function mechanism of our luminesensor. When exposed to blue light, the N-terminal cap of VVD domain will undock and cause VVD domain to dimerize. The dimerization of VVD protein will help the dimerization of N-terminal domains of LexA protien fused to the N-terminal of VVD domain. When helped to dimerize, the NTD of LexA protein will recognize and bind to the SOS box in the promoter of our reporter gene and inhibit transcription initiation.
With the discussion above, we have decided to use DNA binding domain of LexA repressor protein as the physiologically functional domain. With a flexible linker linking the two distinct domains, we now have the panorama of our design of photosensitive fusion protein. We term this VVD-LexA based photosensitive fusion protein ‘Luminesensor’.
Design: Ensuring Orthogonality
Perhaps a careful reader has already noticed a severe defect in our design. That is, by choosing the DNA binding domain of a bacteria endogenous transcription repressor, we must use reporter genes that will also be repressed by endogenous LexA protein. If we do not introduce changes into the DNA binding domain of our luminesensor, the only way to rule out the interference of endogenous LexA would be knocking out the genomic sequence of LexA protein. This will indubitably diminish its applicability in the future, since it will not work in any strain with endogenous LexA remaining.
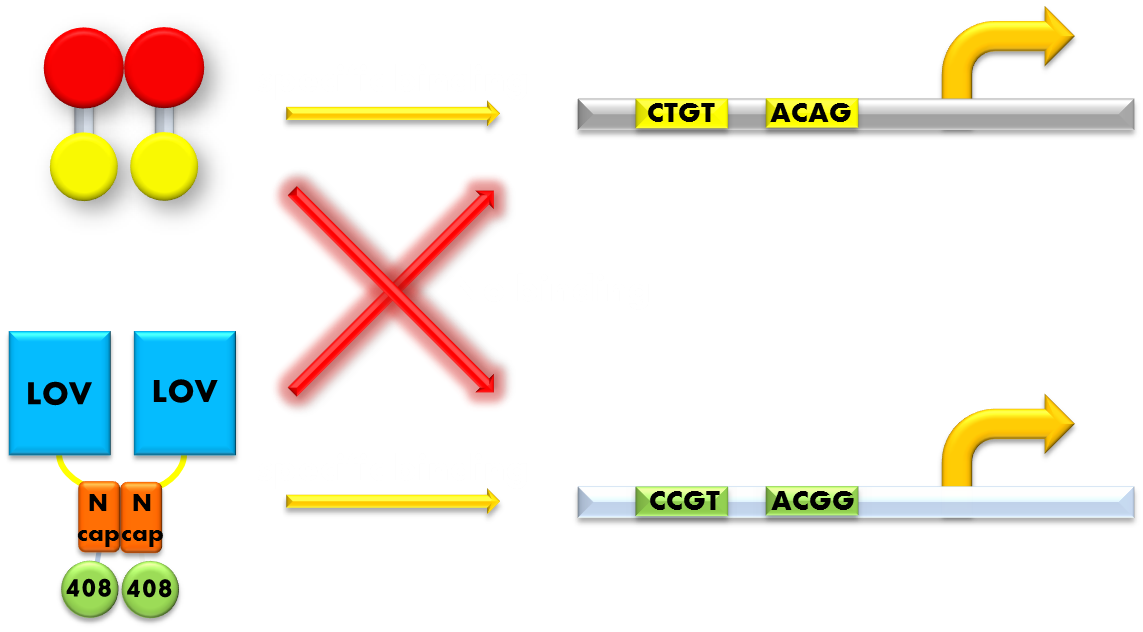
Figure 10. Our solution to the problem of orthogonality. We found that a LexA variant LexA408 will only recognize a symmetrically altered SOS box (CCGT(N)8ACGG), different form the original one (CTGT(N)8ACAG). So we changed the LexA NTD domain of our wild type luminesensor to its 408 form, and changed the SOS box in our reporter gene promoters correspondingly. Now the endogenous LexA protein will no longer bind to the promoters of our reporter gene, while our luminesensor still possessing the light-inducible repression function. Thus we ensured our system's orthogonality to the bacteria SOS system.
So we set out to find a solution to this important problem. The most natural way would be creating a mutated form of LexA so that it will only recognize a target sequence different from the sequence the wild-type LexA would recognize. We gladly found that such a thing actually exists. It is called the LexA408 variant, which carries mutations PA40, NS41 and AS42 and has been shown much higher affinity for a symmetrically altered target sequence CCGT (N)8 ACGG, different from the wild type SOS box CTGT (N)8 ACAG, which is specifically recognized by wild-type LexA. Thus by introducing the 408 mutations into the DNA binding domain of our luminesensor, we can insulate our light-sensitive system from the interference from the genetic context of the host strain. This outstanding character that have ensured the orthogonality of our system to the endogenous SOS system will greatly enhance the compatibility of our module to those synthetic modules that have hitherto been designed and thus add to the promise of future application.
Design: Dynamic Performance Optimization
Though our Luminesensor has outshined many previously designed optogenetic modules in many aspects, space still remains for its improvement. A particularly important matter would be the dynamic performance of our Luminesensor. Signal-background ratio is always a significant judging criterion in synthetic modules. The more digital-like the output may be, the more likely the module would function efficiently in a complex biological system where wanted and unwanted noises are inevitable. Though our Luminesensor has already shown a high background-signal ratio, we still deem dynamic range optimization necessary, since background-signal ratio can never be too high. Another concern would be whether the system is resettable. In our Luminesensor system, this equals to say whether our Luminesensor will be able to decay back fast to its dark state when it is no longer exposed to light. This is particular important when our system is to be applied in a scenario where cell movement is inevitable. For example, imagine we are using liquid medium to culture our Luminesensor containing bacteria and applying to a proportion of the medium blue light to inhibit reporter gene, say GFP, expression to create some sort of pattern, cells that have received blue light will constantly swim out of the illuminated area. If our Luminesensor’s decay half-life is very long, the light-received cells will eventually distribute evenly in those dark area, so no pattern would form. Unfortunately, VVD protein, the photosensor domain we use, has a typical light-to-dark decay half-life of hours. This makes our decay half-life optimization extremely important.
In order to rationally optimize our Luminesensor’s dynamic range and decay half-life, we need to build a dynamic model in the first place to simulate our Luminesensor system and determine which ones of those parameters in the system would be decisive to the two dynamic features we are interested in.
We listed all possible states of the molecules involved in our Luminesensor system and transitions between those states and created an ordinary deferential equation system based on law of mass action to describe the dynamic behavior of our system. After in-silico simulation, we singled out 4 critical parameters in our system that would determine the above two aspects of our system’s dynamic performance.
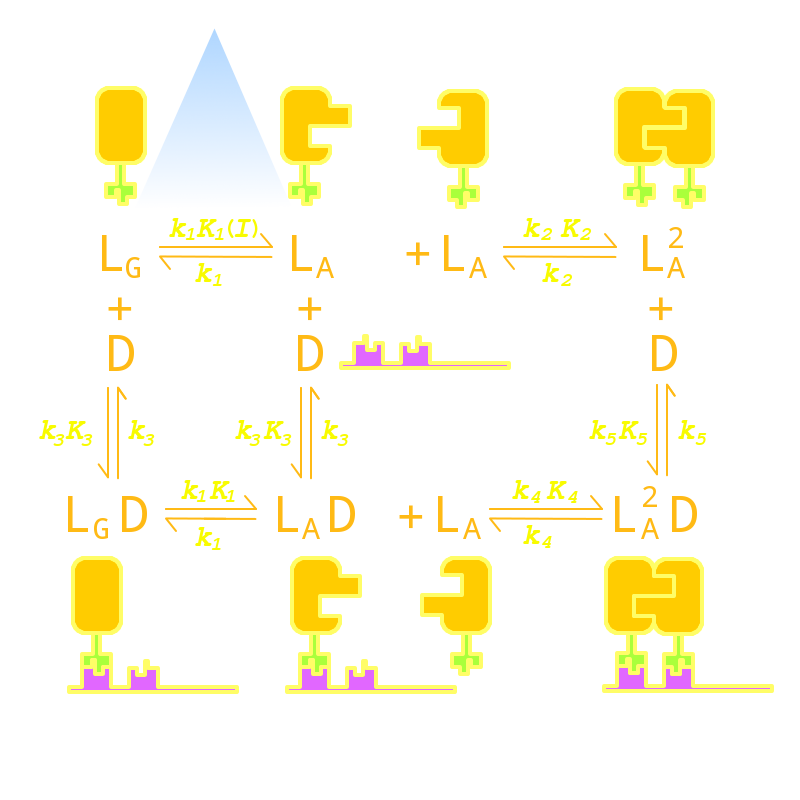
Fig 1. Kinetic Network of our Luminesensor
Two of them will affect the background-signal ratio, namely the VVD protein dimerization equilibrium constant and dimerized Luminesensor’s DNA binding equilibrium constant. This is easy to understand intuitively. Because the tighter the Luminesensors dimerize when illuminated by light, the greater the intracellular concentration of dimerized Luminesensor will be. And the tighter the dimerized Luminesensors bind the target DNA sequence, the greater proportion of target gene will be repressed, thus reducing the background of reporter gene expression under light. The other two parameters are responsible for the decay half-life, namely the rate constant of VVD photosensor domain’s decaying from the light-state back to its dark-state and rate constant of monomer luminesenor’s disassociating form target DNA sequence. This is also easy to understand intuitively. The faster the VVD proteins decay back to its dark-state, the faster the Luminesensor will turn from it dimerized form to its monomer form, which is significantly less efficient in binding target DNA sequence. And the faster the monomer Luminesensors disassociate from the target sequence, the faster the steric hindrance to RNA polymerase created by DNA-bound Luminesensors will be lifted, thus reducing the amount of time required for the system to reset.
Having singled out these four critical parameters, we set out to search through literature to find mutations that would enhance them. We found that there does not seem to exist mutations that would enhance dimerized Luminesensor’s DNA binding equilibrium constant. Rather, those mutations seem to undermine the DNA binding affinity, which would be detrimental to our system. We further reasoned that any mutation that would facilitate the disassociation of monomer Luminesensors from target DNA sequence is also likely to undermine the DNA binding affinity. So we focused on looking for VVD photosensor domain mutations that would promote VVD dimerization and VVD decay. We eventually localized on two specific mutations of VVD photosensor domain: a M135I mutation that will enhance the VVD dimerization, and a I74V mutation in the vicinity of FAD molecule that will enhance the VVD decay rate.
Having determined the plausible mutations, when then introduced mutation M135I, I74V and the combination of the two mutations into our 408 form of Luminesensor. By characterizing the dynamic behavior of the mutated Luminesensor along time course, we discovered that mutation M135I indeed improved the signal-background ratio significantly. However, I74V mutation seems to reduce the signal-background ratio dramatically, rendering our Luminesensor useless. So disregarding of the faster decay half-life it may provide, we have to rule I74V out of our design.
Reference
- 1. Shimizu-Sato, S., Huq, E., Tepperman, J.M., & Quail, P.H.(2002). A light-switchable gene promoter system. Nat. Biotechnol. 20: 1041: 1044
- 2. Wu, Y., Frey, D., Lungu, O.I., Jaehrig, A., Schlichting, I., Kuhlman, B. & Hahn, K.M.(2009). A genetically encoded photoactivatable Rac controls the motility of living cells. Nature, 461: 104: 8
- 3. Levskaya, A., Weiner, O.D., Lim, W.A. & Voigt, C.A.(2009). Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature, 461: 997: 1001
- 4.Möglich, A., Ayers, R.A. & Moffat, K.(2009). Design and Signaling Mechanism of Light-Regulated Histidine Kinases. J. Mol. Biol., 385: 1433: 1444
- 5. Strickland, D., Moffat, K. & Sosnick, T.R.(2008). Light-activated DNA binding in a designed allosteric protein. Proc. Natl Acad. Sci. USA, 105: 10709: 10714
- 6. Ohlendorf, R., Vidavski, R.R., Eldar, A., Moffat, K. & Möglich, A.(2012). From Dusk till Dawn: One-Plasmid Systems for Light-Regulated Gene Expression. J. Mol. Biol., 416: 534: 542
- 7. Toettcher, J.E., Voigt, C.A., Weiner, O.D. & Lim, W.A.(2010). The promise of optogenetics in cell biology: interrogating molecular circuits in space and time. Nat. Methods, 8: 35: 38
- 8. Bacchus, W. & Fussenegger, M.(2011) The use of light for engineered control and reprogramming of cellular functions. Curr. Opin. Biotechnol., 23: 1: 8
- 9. Cole, S.T.(1983) Charaeterisation of the Promoter for the LexA Regulated sulA Gene of Escherichia coli. Mol. Gen. Genet., 189: 400: 404
- 10. Crane, B.R. et al.(2007) Conformational Switching in the Fungal Light Sensor Vivid. Science, 316: 1054: 1057
- 11. Herrou, J. and Crosson, S.(2011) Function, structure and mechanism of bacterial photosensory LOV proteins. Nat. Rev. Microbiol., 9: 713: 723
- 12. Zoltowski, B.D., and Crane, B.R.(2008) Light activation of the LOV protein Vivid generates a rapidly exchanging dimer. Biochemistry, 47: 7012: 7019
- 13. Zoltowski, B.D., Vaccaro, B. & Crane, B.R. Mechanism-based tuning of a LOV domain photoreceptor. Nat. Chem. Biol., 5: 827: 834
- 14. Vaidya, A.T., Chen, C.H., Dunlap, J.C., Loros, J.J., and Crane, B.R.(2011) Structure of a light-activated LOV protein dimer that regulates transcription in Neurospora crassa. Sci. Signal., 4: ra50
- 15. Wang, X., Chen, X. & Yang, Y.(2012) spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods, 9: 266: 269
- 16. Zhang, A.P.P., Pigli, Y.Z & Rice, P.A.(2010) Structure of the LexA–DNA complex and implications for SOS box measurement.Nature, 466: 883: 886
- 17. Butalaa, M., Zgur-Bertokb, D., and Busby, S. J. W.(2009) The bacterial LexA transcriptional repressor. Cell. Mol. Life Sci., 66: 82: 93
- 18. Tabor, J.J., Salis,H.M., Simpson, Z.B., Chevalier,A.A., Levskaya, A., Marcotte, E.A., Voigt, C.A., and Ellington, A.D.(2009) A Synthetic Genetic Edge Detection Program. Cell, 137: 1272: 1281
- 19. Voigt., C.A., et al.(2005) Engineering Escherichia coli to see light. Nature, 438: 441: 442
- 20. Ye, H., Baba, M.D.E., Peng, R., Fussenegger, M.(2011) A Synthetic Optogenetic Transcription Device Enhances Blood-Glucose Homeostasis in Mice. Science, 332: 1565: 1568
 "
"














