Team:SEU O China/Result
From 2012.igem.org
| Line 227: | Line 227: | ||
| + | |||
| + | |||
| + | |||
| + | <html> | ||
| + | <div class="clear"></div> | ||
| + | </div> | ||
| + | |||
| + | <div class="content" id="licontent"> | ||
| + | <h3>Light Sensor</h3><br> | ||
| + | </html> | ||
| + | |||
| + | '''RED LIGHT''' | ||
| + | |||
| + | |||
| + | '''Scheme A''' | ||
| + | |||
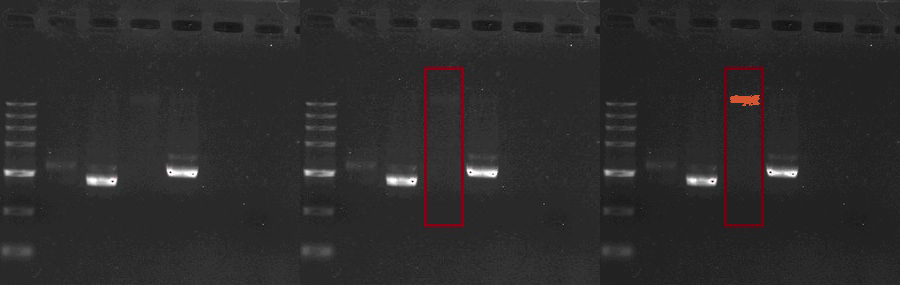
| + | The experiments of light sensor part starts from BBa_M30109, the devised composite that can produce Cph8 and ho1 & pcyA,the enzymes required by convertion of Haem into PCB.The OmpC promoter is also involved in this composite. However, BBa_M30109 is not as available as mentioned on the website. The PCR failed to amplificate the fragment of BBa_M30109. It might be the giant size of this part results in the failure of PCR.Besides the plasmids extracted from the E.coli bacteria can hardly be told from the electrophoretogram (Fig.1). As a result, we decided to choose "scheme B". | ||
| + | [[File:M30109_revised_.JPG|center|570px]] | ||
| + | |||
| + | |||
| + | '''Scheme B''' | ||
| + | |||
| + | We deside to constructe two composites that can produce Cph8, termed c-Cph8 and enzymes we need, named c-PCB, seperately. | ||
| + | |||
| + | C-Cph8 basically contains BBa_R0010, BBa_S03419 and BBa_B0015. The combination of BBa_S03419 and BBa_B0015 first succeeded at the beginning of August. The sequence report suggest the correct sequencing information of both BBa_S03419 and BBa_B0015. Sadly, problems came to our experiments when we tried to recover the BBa_R0010’s seed conserved by ourselves. The seed we conserved on that day was somehow polluted so that we failed to extract the correct and available plasmids. So we just finished the construction of BBa_S03419 and BBa_B0015, termed BBa_S05053, which contained an RBS, BBa_I15008 and the terminator BBa_B0015. | ||
| + | |||
| + | C-PCB is expected to produce the two vital enzymes, ho1 and pcyA that catalyze the procedure of conversion of haem into PCB. We planned to combine BBa_J13002, BBa_I15008, BBa_K081024 and a reporter protein(mRFP) together. When we finished the combination of BBa_K081024 and BBa_I13507(mRFP), we were astonished to find out mRFP was expressed without Cph8. Meanwhile, the combination of BBa_I15008 and BBa_J13002 failed. The good news is that the sequencing report confirmed the sequences of BBa_13002, BBa_K081024 and BBa_I13507 were seperately right. Unfortunately, the inversion of BBa_I15008 succeeded but we lost the correct seed due to an accident. As a result, we finished the combination of BBa_K081024 and BBa_I13507, termed BBa_S05054 automatically by partsregistry.org. | ||
| + | [[File:https://2012.igem.org/File:SeuoRFP.jpg|center]] | ||
| + | |||
| + | |||
| + | '''Discussion''' | ||
| + | |||
| + | The expression of mRFP in Scheme B suggest that the OmpC in BBa_K081024 was somehow activated. Further work was conducted to figure out why the OmpC seemed to present a function of a continuous promoter. After blasting the sequence of Cph1 at NIBC (http://www.ncbi.nlm.nih.gov), we discovered that the BL21(DE3) strain has endogenous EnvZ, which is the key part of Cph1that can phosphorylate the OmpR and cause the activation of OmpC followed by the expression of downstream genes, BBa_I13507. Hence, once the strain has endogenous EnvZ, BBa_K081024 & BBa_I13507, also known as BBa_S05054 expresses mRFP without Cph8 not to mention the regulation of red light. | ||
| + | |||
| + | Now it is confirmed that as long as equipped with a promoter,RBS and BBa_I15008, the OmpC promoter in BBa_S05054 can work normally in a endogenous-EnvZ-free system. Besides, BBa_S05054 can also be utilized to test the exisitence of endogenous EnvZ. | ||
| + | |||
| + | |||
| + | '''BLUE LIGHT''' | ||
| + | |||
| + | Blue light system is much simple compared with red light system, since it involves just one kit, BBa_K238013. It eventually failed in the procedures of combination with other part of our project. | ||
Revision as of 09:47, 26 September 2012


Results
Biobricks
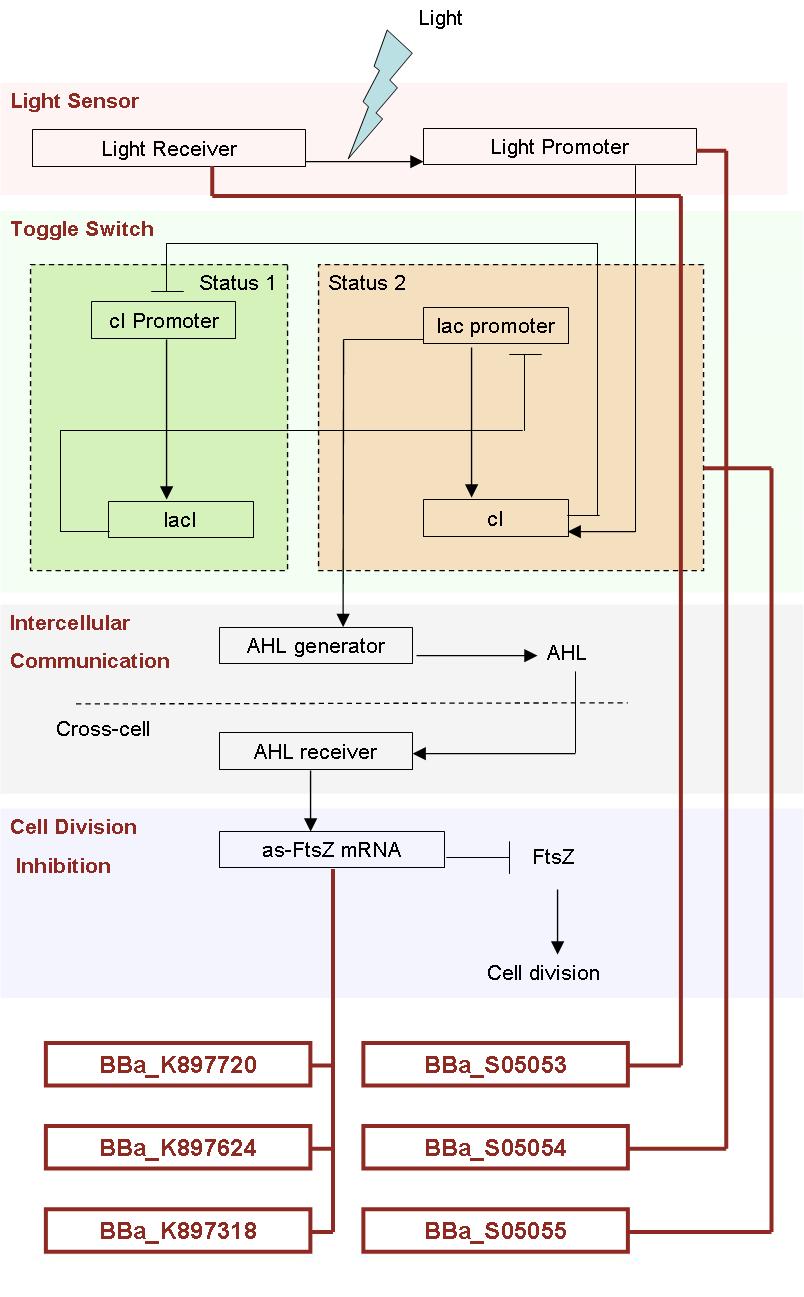
How our system works:
Our Favorite New Parts:
| Name: | TBY | ID: | [http://partsregistry.org/Part:BBa_K897720 BBa_K897720] | |
| Type: | RNA | Length: | 382bp | |
| Description: | This part encodes a fragment of antisense FtsZ RNA protected by paired termini structure under T7 promoter. Inhibiting the expression of FtsZ, it can strongly repress the cell division in E.coli. | |||
*TBY means "Te Bie Yuan", or "very round" in English...
Other Parts We like:
| Name: | FtsZ_B0015 | ID: | [http://partsregistry.org/Part:BBa_K897624 BBa_K897624] | |
| Type: | RNA | Length: | 283bp | |
| Description: | This part encodes a fragment of antisense FtsZ RNA combined with a terminator. It can repress the cell division in E.coli when used with a promoter. | |||
.
| Name: | Paired Termini | ID: | [http://partsregistry.org/Part:BBa_K897318 BBa_K897318] | |
| Type: | Other | Length: | 146bp | |
| Description: | This part encodes a paired termini with a MCS in the middle of it. By insert target antisense RNA into the MCS, it can protect the antisense RNA from degradation and enhance the efficiency or antisense RNA silencing. | |||
.
| Name: | S03419_B0015 | ID: | [http://partsregistry.org/Part:BBa_S05053 BBa_S05053] | |
| Type: | Intermediate | Length: | 2418bp | |
| Description: | This part is the combination of S03419 and B0015, a part of light sensor system. Once equipped with a promoter, BBa_S05053 can produce the Cph8,the key menbrane-bound protein of red light sensor. | |||
.
| Name: | K081024_I13507 | ID: | [http://partsregistry.org/Part:BBa_S05054 BBa_S05054] | |
| Type: | Intermediate | Length: | 1828bp | |
| Description: | BBa_S05054 is an intermediate constructed by the conjunction of K081024 and I13507. As a part of light sensor, it contains the PcyA coding sequence(I15009), the OmpR-controlled promoter(ROO82) and the mRFP reporter(I13507. With a combination of J13002 and I15008,this part can produce the enzymes, whicn can catalyzed the two procedures required by conversion of haem into PCB,and offer the OmpC promoter. | |||
.
| Name: | R0010_P0451 | ID: | [http://partsregistry.org/Part:BBa_S05055 BBa_S05055] | |
| Type: | Intermediate | Length: | 1138bp | |
| Description: | This part is the pre-half of the collins toggle switch, which include a lac promoter (ROO1O) and cI coding part(P0451). It can be used as a toggle switch when combined with R0051_I732820. | |||
.
Data For Pre-existing Parts:
[http://partsregistry.org/Part:BBa_M30109:Experience BBa_M30109 Experience:]
- BBa_M30109 seems not as available as mentioned on the website. We tried PCR and extraction of plasmids successively but both of them failed. It might be the size that resulted in the instability of those test procedures since BBa_M30109 has an astonishing size of 4333bp: with such a giant fragment it may lead to multiple mistakes in replication.
[http://partsregistry.org/Part:BBa_R0051:Experience BBa_R0051 Experience:]
- BBa_R0051 does not work well due to its rather small size. We transformed it for three times, and none of them succeed. We designed primers to PCR it out from K091230 successfully, but failed to perform the digestion since it is too small to extract from gel. It may work better if adding some nonsense sequence before the promoter.
[http://partsregistry.org/Part:BBa_J5526:Experience BBa_J5526 Experience:]
- We tried to transform BBa_J5526 several times. It grows well, but never turns red.
Division Inhibition
The antisense FtsZ, protected by hair-pin structure and termed TBY or BBa_897720, is regulated by T7 promoter and regarded as the core part of our project. The antisense FtsZ sequence mainly aims at silenting the FtsZ gene and control the self-renewal and differentiation of E.coli. Since the antisense FtsZ gene in BBa_897720 is regulated by the T7 promoter that is specifically combined with T7 RNA polymerase, a endogenous protein in BL21(DE3) strain under the control of the lac UV5 promoter inducible by IPTG, the number of colonies is expected to decrease with the an increase in IPTG. The confirmatory experiment below verifys the validity of TBY and our hypothesis.
After integrating the plasmid with an insert of BBa_897720 into BL21(DE3) strain, we cultivated those bacteria on the surface of solid LB medium in 4 dfferent IPTG levels. The conditions was strictly controlled at the temperature of 37 ℃ and it last for 10 hours. The controls aims to rule out the possibility of damaged to the bacteria from IPTG. Details are illustrated below:
| IPTG(0.0mM) | IPTG(0.5mM) | IPTG(1.0mM) | IPTG(2.0mM) | |
| TBY *0.01 | 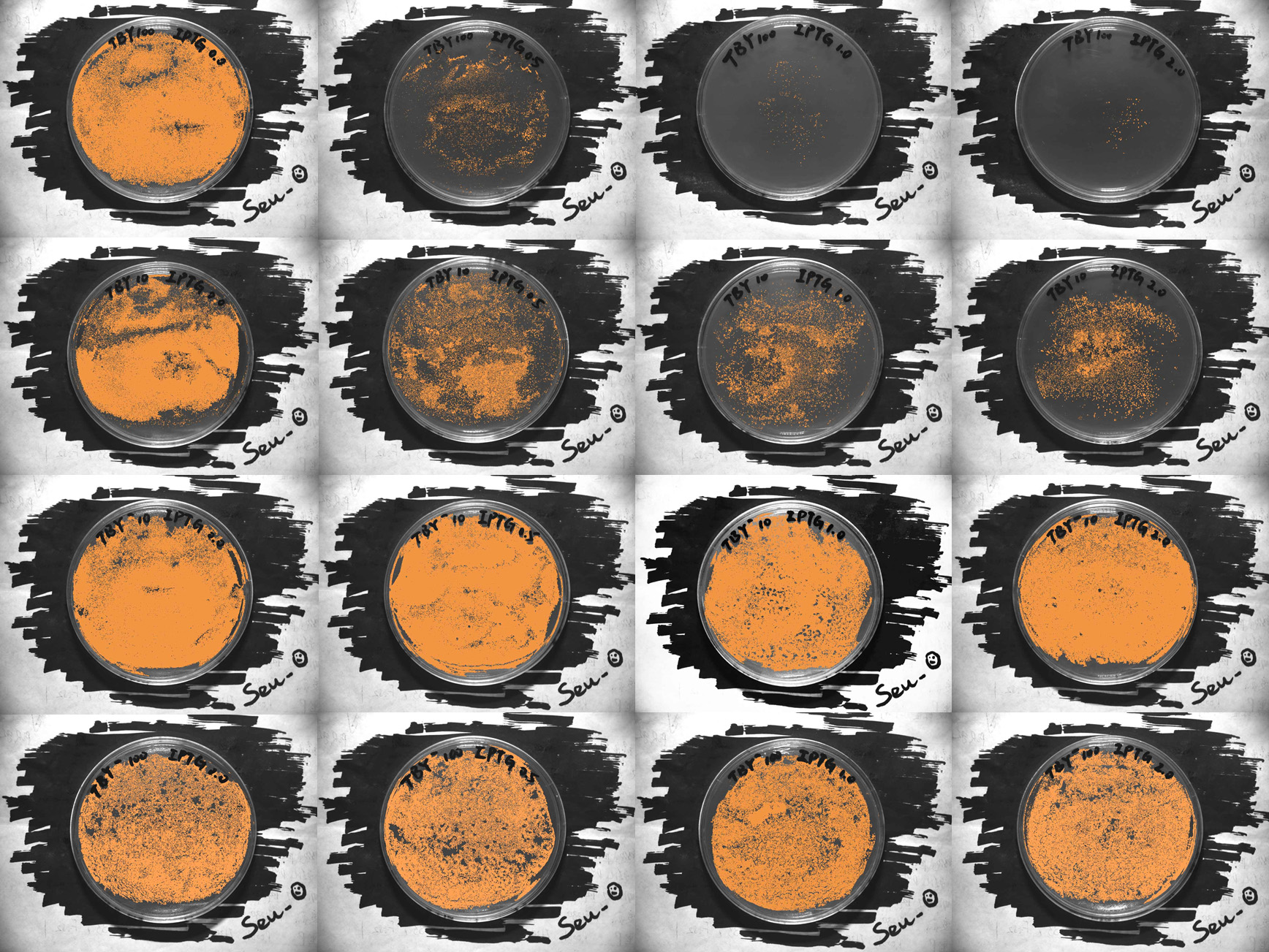
| |||
| TBY *0.1 | ||||
| TBY- *0.1 | ||||
| TBY- *0.01 | ||||
- TBY represents the BL21(DE3) bacteria carring plasmids with an insert of BBa_K897720. TBY- means that the BL21(DE3) bacteria carried the plasmids as same as the plasmids of TBY but without the insert of BBa_K8977720.
- TBY(-) are diluted tenfold and hundredfold separately, marked as “*0.1” and “*0.01”.
- The number in the parenthesis following the “IPTG” means the concentration of IPTG.
- For original full-size picture, see here.
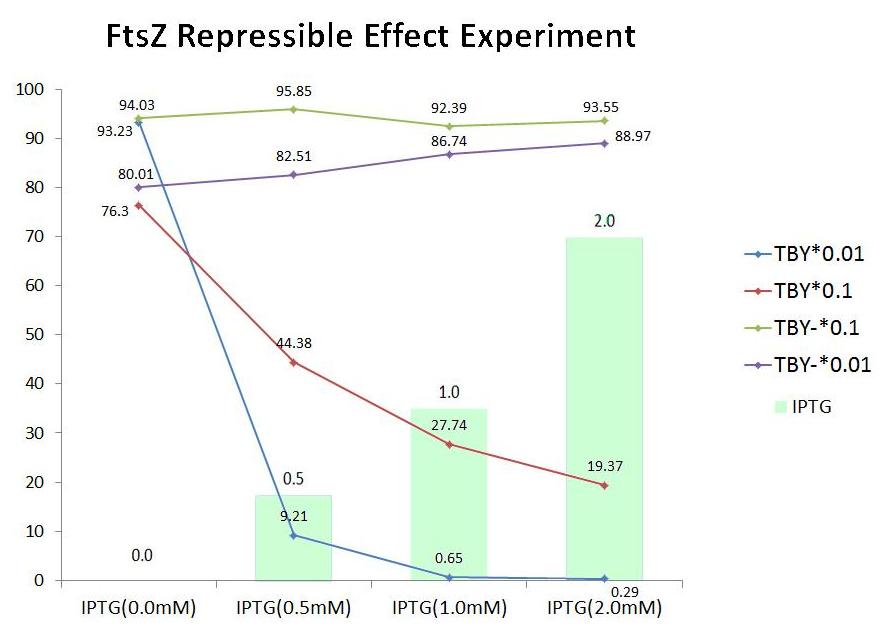
In order to analyse the repressible effect of FtsZ, we calculate the colony coverage of every sample. Details are illustrated in the Model Part, and the results of analysis are shown below.
| IPTG(0.0mM) | IPTG(0.5mM) | IPTG(1.0mM) | IPTG(2.0mM) | |
| TBY *0.01 | 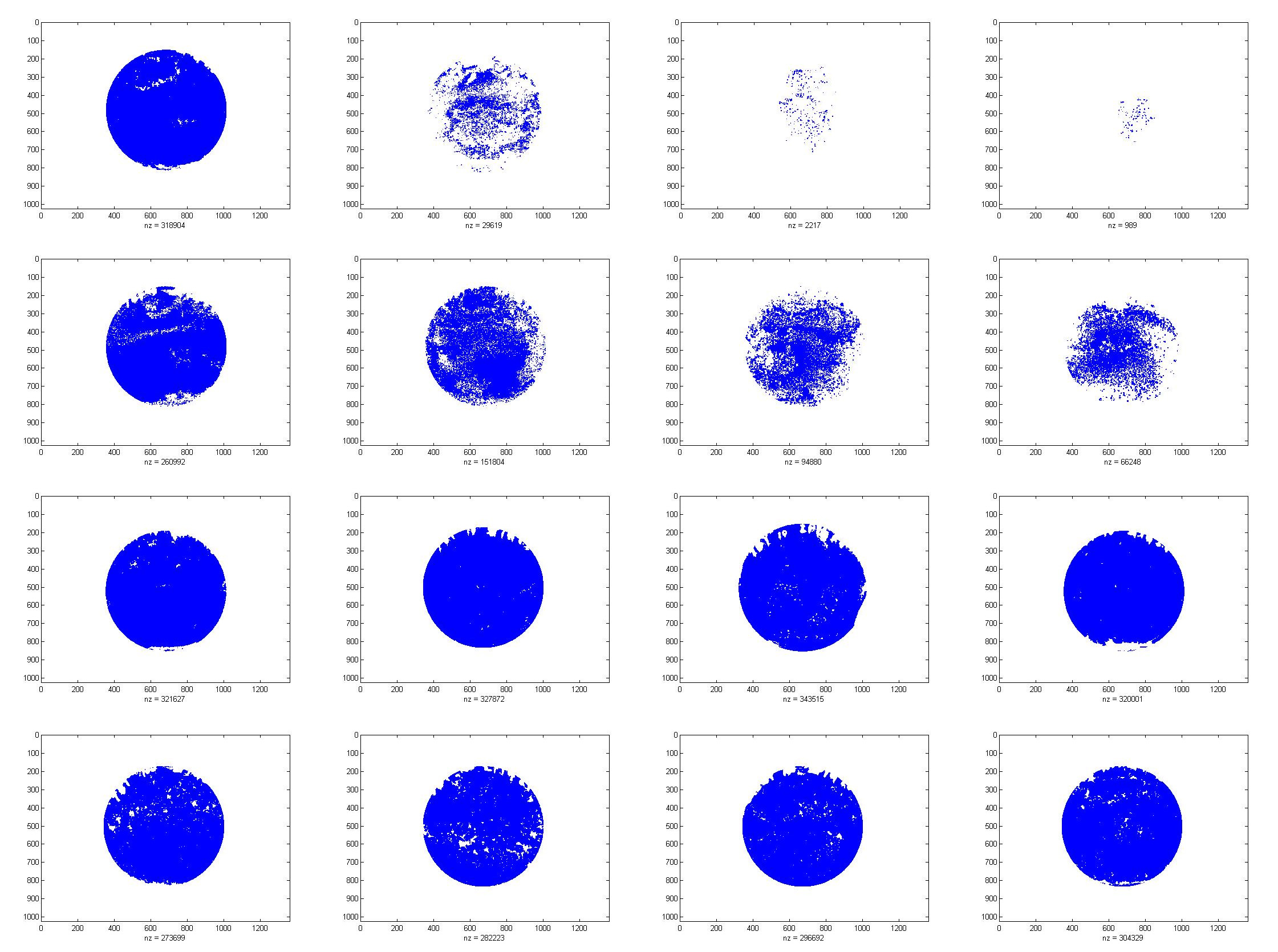
| |||
| TBY *0.1 | ||||
| TBY- *0.1 | ||||
| TBY- *0.01 | ||||
- Colony coverage Map
| IPTG(0.0mM) | IPTG(0.5mM) | IPTG(1.0mM) | IPTG(2.0mM) | |
| TBY*0.01 | 93.23 | 9.21 | 0.65 | 0.29 |
| TBY*0.1 | 76.30 | 44.38 | 27.74 | 19.37 |
| TBY-*0.1 | 94.03 | 95.85 | 92.39 | 93.55 |
| TBY-*0.01 | 80.01 | 82.51 | 86.74 | 88.97 |
- Percentage of colony coverage
- FtsZ repressible effect
Both of the red line and the blue line in the illustration show that the colony coverage decreases while the level of IPTG increases, which suggests that the growth of colony is somehow repressed. Moreover, the yellow line reveals that the IPTG has little side effect on colony.
As a result, FtsZ truly represses the growth of bacteria and also provide experimental evidence for our project scheme.
Light Sensor
RED LIGHT
Scheme A
The experiments of light sensor part starts from BBa_M30109, the devised composite that can produce Cph8 and ho1 & pcyA,the enzymes required by convertion of Haem into PCB.The OmpC promoter is also involved in this composite. However, BBa_M30109 is not as available as mentioned on the website. The PCR failed to amplificate the fragment of BBa_M30109. It might be the giant size of this part results in the failure of PCR.Besides the plasmids extracted from the E.coli bacteria can hardly be told from the electrophoretogram (Fig.1). As a result, we decided to choose "scheme B".
Scheme B
We deside to constructe two composites that can produce Cph8, termed c-Cph8 and enzymes we need, named c-PCB, seperately.
C-Cph8 basically contains BBa_R0010, BBa_S03419 and BBa_B0015. The combination of BBa_S03419 and BBa_B0015 first succeeded at the beginning of August. The sequence report suggest the correct sequencing information of both BBa_S03419 and BBa_B0015. Sadly, problems came to our experiments when we tried to recover the BBa_R0010’s seed conserved by ourselves. The seed we conserved on that day was somehow polluted so that we failed to extract the correct and available plasmids. So we just finished the construction of BBa_S03419 and BBa_B0015, termed BBa_S05053, which contained an RBS, BBa_I15008 and the terminator BBa_B0015.
C-PCB is expected to produce the two vital enzymes, ho1 and pcyA that catalyze the procedure of conversion of haem into PCB. We planned to combine BBa_J13002, BBa_I15008, BBa_K081024 and a reporter protein(mRFP) together. When we finished the combination of BBa_K081024 and BBa_I13507(mRFP), we were astonished to find out mRFP was expressed without Cph8. Meanwhile, the combination of BBa_I15008 and BBa_J13002 failed. The good news is that the sequencing report confirmed the sequences of BBa_13002, BBa_K081024 and BBa_I13507 were seperately right. Unfortunately, the inversion of BBa_I15008 succeeded but we lost the correct seed due to an accident. As a result, we finished the combination of BBa_K081024 and BBa_I13507, termed BBa_S05054 automatically by partsregistry.org.
Discussion
The expression of mRFP in Scheme B suggest that the OmpC in BBa_K081024 was somehow activated. Further work was conducted to figure out why the OmpC seemed to present a function of a continuous promoter. After blasting the sequence of Cph1 at NIBC (http://www.ncbi.nlm.nih.gov), we discovered that the BL21(DE3) strain has endogenous EnvZ, which is the key part of Cph1that can phosphorylate the OmpR and cause the activation of OmpC followed by the expression of downstream genes, BBa_I13507. Hence, once the strain has endogenous EnvZ, BBa_K081024 & BBa_I13507, also known as BBa_S05054 expresses mRFP without Cph8 not to mention the regulation of red light.
Now it is confirmed that as long as equipped with a promoter,RBS and BBa_I15008, the OmpC promoter in BBa_S05054 can work normally in a endogenous-EnvZ-free system. Besides, BBa_S05054 can also be utilized to test the exisitence of endogenous EnvZ.
BLUE LIGHT
Blue light system is much simple compared with red light system, since it involves just one kit, BBa_K238013. It eventually failed in the procedures of combination with other part of our project.
Toggle switch
The toggle switch part of experiment is not very successful.
We have constructed the lac-cI part of the toggle switch, but the pcI-lacI part is failed. The most problem happened around the cI promoter, R0051, which is only 49bp in length. After the failure of several transformation and depletion of the distributed kit, we try to PCR this part out from K091230, a cI promoted mRFP reporter.
Though the PCR works well and we actually get the target fragment, we failed to perform follow experiment because we cannot extract DNA from the enzyme digestion product since the 49bp size is beyond the ability of our Gel Extraction Kit.
After that, we tried to add some nonsense sequence before K091230, and PCR the sequence and R0051 out, but finally did not make it before the deadline.
Judging form
Attribution

 "
"