Team:Bielefeld-Germany/Labjournal/week15
From 2012.igem.org
(Difference between revisions)
(→weekly meeting) |
(→weekly meeting) |
||
| Line 58: | Line 58: | ||
| - | <span style="color:#FFFFFF"> 8====& | + | <span style="color:#FFFFFF"> 8====∋~ </span> |
===Monday August 6th=== | ===Monday August 6th=== | ||
Revision as of 15:08, 25 September 2012
Contents |
Week 15 (08/06 - 08/12/12)
weekly meeting
8====∋~
Monday August 6th
- Team Site Directed Mutagenesis:
- Plasmid-isolation of two more ecol-g2307a colonies.
- Team Cellulose Binding Domain:
- Plated one colony of <partinfo>BBa_I13522</partinfo> on selection-agar.
- Dephosphorylation of pSB1C3 Backbone.
- Ligation of [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] with the pSB1C3-Backbone and [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] with the pSB1C3-Backbone.
- Transformed both in KRX and plated on CM-selection-agar.
- Team Cloning of Bacterial Laccases:
- No positive colonies after transformation of our assemblies from August 1st but we realized that the primers we used for making the promoter parts can’t ligate with our backbone because the primers are dephosphorylated and the plasmid backbone is dephosphorylated, too. Much effort in a mission which can't work but at least we know now why it doesn't work.
- Picking positive colonies from transformation of ecol and ecol_HIS in pSB1C3 for plasmid isolation.
- Team Fungal and Plant Laccases: Plating positive colonies from cloning of tvel5 in pSb1C3 backbone.
- Team Immobilization:
- CPC beads have just arrived! We couldn’t wait any longer to start our new experiments. Since the amount of beads used and the incubation period varied from one paper to another (see Literature), we decided to find out ourselves the most convenient conditions for our laccases. We started with different bead concentrations: 0.01, 0.02, 0.04, 0.06, and 0.08 mg/mL. The beads were first immersed in 2.5% glutaraldehyde and incubated for 2 hours under light vacuum in order to allow as much beads’ surface area to be coated with aldehyde groups, which crosslink the laccases to the beads. After that, the beads were washed 3 times with Britton-Robinson Buffer (link) and then immersed in 1mg/mL laccases from TVEL0 and placed on a rotator at 4°C for 18h and 36h respectively. For further information see Protocol.
Tuesday August 7th
- Team Cloning of Bacterial Laccases:
- Plasmid isolation and control restriction of ecol and ecol_HIS in pSB1C3 showed correct fragment sizes in agarose gel. So we did a digest for prefix insertion of the new T7 promoter.
- Team Site Directed Mutagenesis:
- Digestion of the two ecol-g2307a-mutants showed that one has lost the restriction-site. Prepared that one for sequencing.
- Team Cellulose Binding Domain:
- Plasmid-isolation of <partinfo>Bba_I13522</partinfo>.
- PCR with <partinfo>BBa_I13522</partinfo> as template and GFP_Freiburg and GFP_His6_compl-primers and clean-up through agarose-gel.
- Restriktion of Linear Backbone, [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] and [http://partsregistry.org/Part:BBa_K863121 GFP_His] with EcoRI and PstI.
- Ligation of Linear Backbone with [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] and with [http://partsregistry.org/Part:BBa_K863121 GFP_His], respectively.
- Transformation of all that stuff and plating on selection-agar.
- Team Immobilization:
- The 18h samples (see above: Monday August 6th/Team Immobilization) were ready for further analysis. The supernatants were gathered to be tested for laccase activity. The beads were then washed with Britton-Robinson Buffer (link), then with 0.5M NaCl solution in order to wash away all noncovalently bound laccases and again twice with the buffer. Subsequently, the beads were immersed in 2.5 mg/mL glycine for 18h at 4°C.
Wednesday August 8th
- Team Cloning of Bacterial Laccases:
- We dephosphorylated the digested the plasmids from day before and phosphorylated the promoter parts. After that we ligated the two parts and transformated the products into KRX electrocompetent cells.
- Team Fungal and Plant Laccases: Plasmid isolation of tvel5 laccase in pSB1C3 backbone.
- Team Site Directed Mutagenesis:
- Made PCRs on tvel-t143g-mutants with Tv_lac10.P.FW and Tv_lac10.S.RV primers, with products of 1.6 kbp when there should be about 4.0 kbp.
- Team Cellulose Binding Domain:
- Picked one not-red colony of [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)], [http://partsregistry.org/Part:BBa_K863112 CBDclos] and [http://partsregistry.org/Part:BBa_K863121 GFP_His] and plated it on seletion-agar
- Restriktion of [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)]-PCR-product with AgeI and EcoRI with DpnI
- Restriktion of [http://partsregistry.org/Part:BBa_K863121 GFP_His]-PCR-product with NgoMIV and PstI and DpnI
- Freiburg-Assembly with [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] and [http://partsregistry.org/Part:BBa_K863121 GFP_His] on the linearized pSB1C3 Backbone followed by plating on selection-agar.
- Team Activity Tests: Today was an exciting day for us. We got samples from Team Immobilization for activity measurements. They were trying to analyze which concentration of beads is a good choice for immobilizing the laccases. The exciting part was to check all the solvents they are using for possible reaction with ABTS during measurements. Check Team Immobilization´s Labjournal and protocols for the output and further information.
- Team Immobilization:
- The 18h samples (see above: Monday August 6th/Team Immobilization and Tuesday August 7th/Team Immobilization) were washed with Britton-Robinson Buffer (link), then with 0.5M NaCl solution and again twice with the buffer. The 36h samples were treated similarly as the 18h samples (see above: Tuesday August 7th/Team Immobilization). The supernatants were delivered to the “Activity Test” team to measure the activity of the non-bound laccases. Based on the results, we would be able to figure out the most convenient bead concentration and incubation period. After comparing the results, we found out that the activity of the supernatant after 36h was less than after 18h, which indicated that 36h incubation is more convenient. On the other hand, the activity also decreased upon increasing the beads’ concentration without reaching a saturation level (see Fig.). Therefore, we decided to carry out another experiment with higher beads concentration.
Thursday August 9th
- Team Fungal and Plant Laccases:
- Again: Ligation of tvel35 in pSB1C3 backbone.
- Plasmid isolation of tvel5 + pSb1C3 and control digest with NotI. For one of the two plasmids the digest looks good. We sent this plasmid for sequencing.
- Team Cloning of Bacterial Laccases:
- We sent ecol and ecol_His (both in pSB1C3) for sequencing.
- We did again a gradient PCR for the Streptomyces but this time without S. tuebingen because the DNA we got was empty. The results can be seen on the following picture.

Agarosegel from the Streptomyces gradient PCR. 1-12: S. goettingen in Sequence with following Annealing temp. 54.9°C, 55.1°C, 55.8°C, 56.8°C, 58.1°C, 59.5°C, 61.0°C, 62.4°C, 63.7°C, 64.8°C, 65.6°C, 66°C. 13-22: S. tuebingen in Sequence with following Annealing temp. 54.9°C, 55.1°C, 55.8°C, 56.8°C, 58.1°C, 59.5°C, 61.0°C, 62.4°C, 63.7°C, 64.8°C
- Team Cultivation & Purification:
- We got E. coli KRX with our own BioBrick [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] in pSB1C3 like the rest.
- We discussed if we did not produce anything because of the toxicity of our protein, which may reduce the plasmid stability. We searched for maximal used concentration of chloramphenicol and found out that concentrations up to 170 µg/mL were used. Therefore we decided to start a new flask cultivation with concentrations of chloramphenicol varying in 5 steps from 20 µg/mL to 170 µg/mL. Today we made the precultures of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] as well as [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and ?[http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010]?. We used E. coli KRX as negative and [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] as positive control.
- Team Site Directed Mutagenesis:
- Sequencing results arrived:
- One bpul-plasmid is positive on both mutations
- All Xccl-mutants are negative (there even was a part of 900 bp gone missing!)
- plated six more colonies of the xccl-g3633c for plasmid-isolation
- PCR with the original tvel10-plasmid and the Prefix/Suffix-primers, showed no product at all.
- Sequencing results arrived:
- Team Cellulose Binding Domain:
- Plasmid-isolation of [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)], [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] and [http://partsregistry.org/Part:BBa_K863121 GFP_His].
- Transformated the assembly again with more of volume of the assembly-CBDcex-GFP_His-mix.
Friday August 10th
- Team Cloning of Bacterial Laccases:
- Again we did the digest of our new T7 promoter part and the ligation in pSB1C3 backbone with ecol PCR products with and without HIS tag. After that we transformed the ligations in pSB1C3. Additionally we did the same with promoter J23110 instead of T7 promoter.
- We did PCR on [http://partsregistry.org/Part:BBa_K863020 BBa_K863020] with the primers B.halo_FW and B.halo_RV for cloning the gene in pSB1C3 backbone without promoter and HIS tag.
- We ligated the digested pSB1C3 plasmids with ecol and ecol_HIS with the new pT7 promoter and pSB1C3 backbone and transformed the approaches in KRX.
- Team Cultivation & Purification:
- Flask cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010] to test various concentrations of chloramphenicol. We used E. coli KRX as negative and E. coli KRX with [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] as positive control.
- Settings: 100 mL flasks without baffles, final volume: 10 mL, autoinduction medium with 20/ 57,5/ 95/ 132,5/ 170 µg/mL chloramphenicol, 37 °C, 120 rpm, double determination.
- Team Site Directed Mutagenesis:
- Plasmid-isolation xccl-g3633c-colonies and test-digestion showed no positive mutated plasmids.
- New pfu-PCR on the xccl-plasmid with xccl-g3633c primer-mix
- Team Cellulose Binding Domain:
- Picked [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His] assembly colony and plated it on CM-selection-agar for plasmid-isolation.
- Restricted [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)]with EcoRI and AgeI
- Digested insert did not appear in the
- Colony-PCR of [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)].
- Picked and plated several colonies of the [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His]-assembly and plated the on CM-Agar.
Saturday August 11th
- Team Cloning of Bacterial Laccases:
- Colony PCRs showed positive colonies for ecol(J23110) so we plated this colonies on new plates.
- We did the PCRs of the laccase genes ecol, bpul, bhal and lthl again. We used the …_FW / …_RV primers and the …_FW / …_RV_HIS primers of the different genes. Digestion of this PCR products and ligation with pT7 or promoter J23110 and the pSB1C3 plasmid backbone.
- Team Site Directed Mutagenesis:
- Agarose-gel-electrophoresis of xccl-PCR (yesterday) showed bands at 3.2 kbp and over 12 kbp but not at 4 kbp as it should be.
- Team Cellulose Binding Domain:
- A few of the plated pSB1C3+CDBcex+GFP-assembly colonies have grown
- Colony-PCR [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)]-colonies showed no positive clones.
Sunday August 12th
- Team Cloning of Bacterial Laccases:
- PCR cleanup of the PCR products from the day before.
- We did control digest and got bands for ecol with J23110 promoter and with the new pT7. So we sent this plasmids for sequencing.
- Team Site Directed Mutagenesis:
- pfu-PCR with xccl-plasmid and xccl-g2247c primers resulted in no product.
- PCR with pre- and suffix primers for tvel10 with original Topo-plasmid as template with increasing temperature at each step (0.1°C) resulted in no product.
- Team Cellulose Binding Domain:
- Colony-PCR for [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)], [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] and [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His].
- resulted in two positve [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)]-colonies.
- Did another colony-pcr on [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] and [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His] that brought up nine positive clones of [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)].
- Colony-PCR for [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)], [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] and [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His].
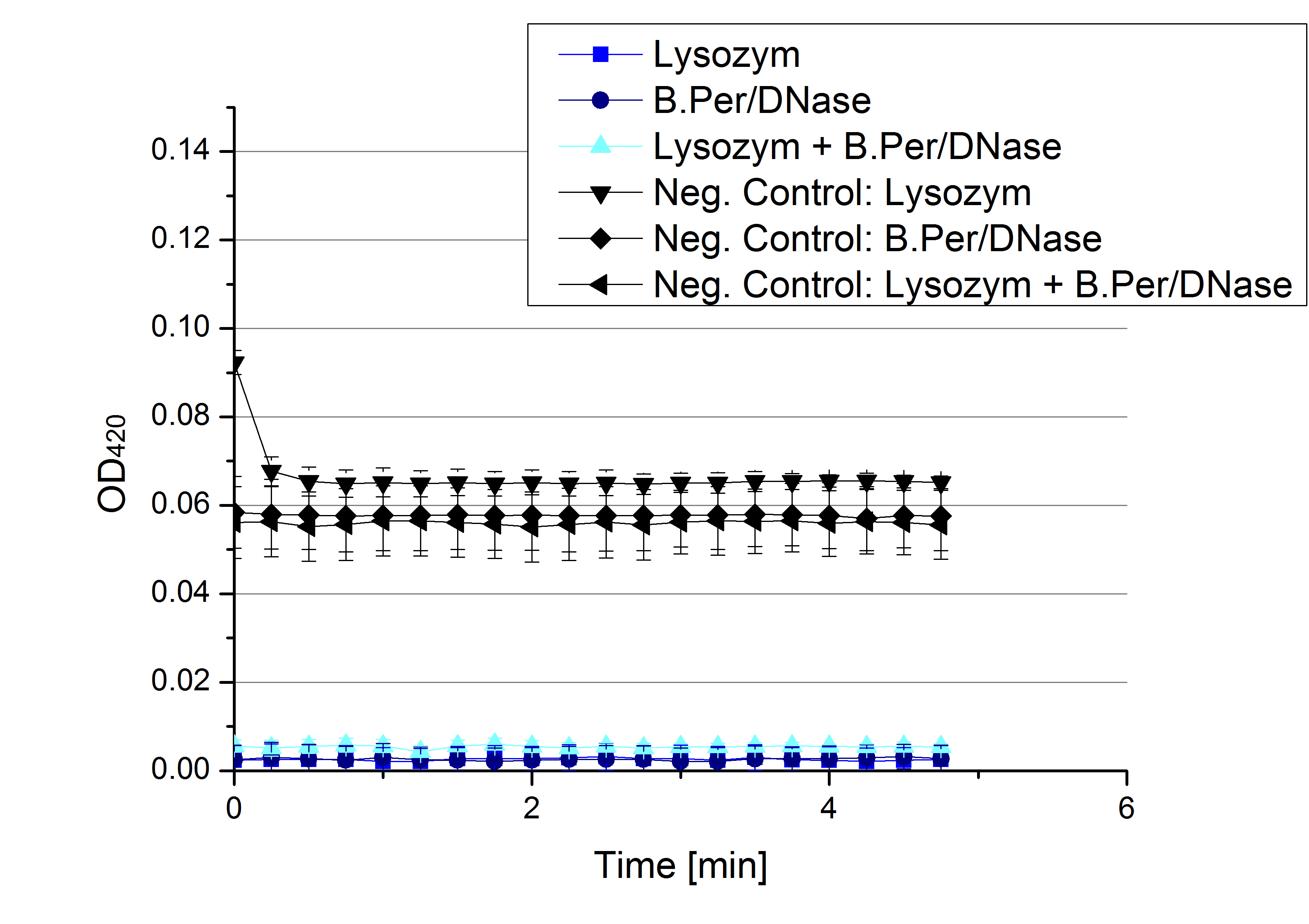
- Team Activity Tests: Today was another day for finding the mistake. Team Cultivation used different methods the for lysis of E.coli KRX cell extract to check whether our [http://partsregistry.org/Part:BBa_K863005 ECOL] laccases and thus their activity might me damaged during lysis. Compared were an enzymatic lysis via lysozyme and a chemical lysis via B.Per/DNase, also both ways were combined in the third sample We are sorry to tell that no activity was seen.
| 55px | | | | | | | | | | |
 "
"