Team:USP-UNESP-Brazil/Plasmid Plug n Play/Results
From 2012.igem.org
Andresochoa (Talk | contribs) |
Andresochoa (Talk | contribs) |
||
| Line 11: | Line 11: | ||
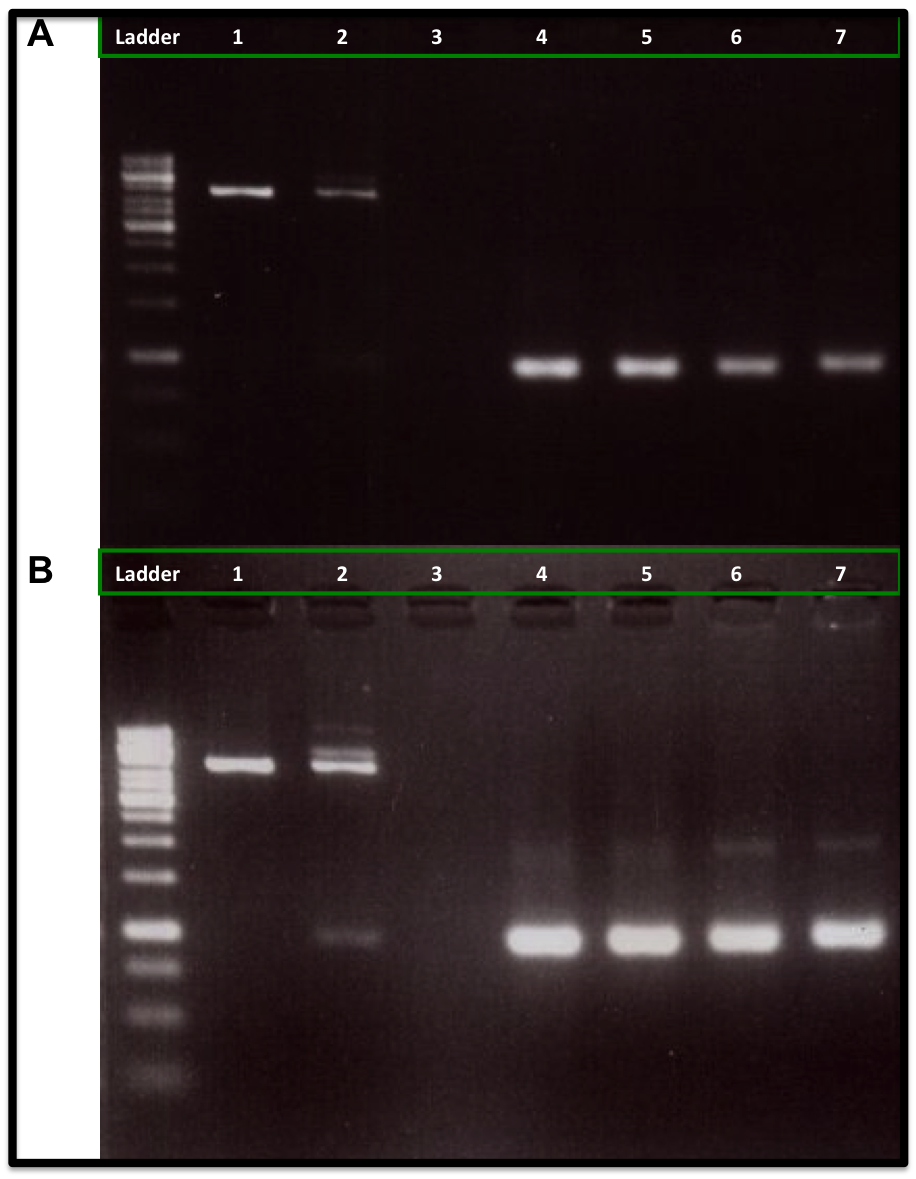
The conclusion was that we can use this loxP-lox66 mechanism in our design and we will need at least 5U of Cre recombinase for any ''in vitro'' experiments. | The conclusion was that we can use this loxP-lox66 mechanism in our design and we will need at least 5U of Cre recombinase for any ''in vitro'' experiments. | ||
| - | {{:Team:USP-UNESP-Brazil/Templates/RImage | image=Reulst1.png | caption= 1kb ladder, 1) Control substrate from the NEB kit without Cre recombinase, 2) Control substrate from the NEB kit with 1U Cre, 3) Empty, 4) Kanamycin resistance gene, 5) Kanamycin resistance gene with 1U Cre, 6) Kanamycin resistance gene with 5U Cre, 7) Kanamycin resistance gene with 10U Cre. | size= | + | {{:Team:USP-UNESP-Brazil/Templates/RImage | image=Reulst1.png | caption= 1kb ladder, 1) Control substrate from the NEB kit without Cre recombinase, 2) Control substrate from the NEB kit with 1U Cre, 3) Empty, 4) Kanamycin resistance gene, 5) Kanamycin resistance gene with 1U Cre, 6) Kanamycin resistance gene with 5U Cre, 7) Kanamycin resistance gene with 10U Cre. | size=600px }} |
| + | |||
Revision as of 03:15, 25 September 2012
 Introduction
Introduction Project Overview
Project Overview Plasmid Plug&Play
Plasmid Plug&Play Associative Memory
Associative MemoryNetwork
 Extras
ExtrasIn vitro assay
This experiments was made to test if the loxP and the lox66 were working properly, it also allowed us to measure the Cre concentration needed in an in vitro recombination reaction. The particularity of lox66 is that it has an altered sequence at the end of it's left arm compared to loxP. This sequence variation reduces affinity of the Cre recombinase for the arm.
Based on the report made by the igem2010 UT-Tokyo team and some papers, the lox66 (BBa_I718016) from the registry was wrong, it had a gg instead a cg in its left arm. This part was corrected by the iGEM11_Tokyo_Tech team (BBa_K649206) and by the iGEM11_WITS_CSIR_SA team (BBa_K537019), but no DNA was available in the registry, so we decided to synthesized it and test it, using the proper sequence described by http://www.ncbi.nlm.nih.gov/pmc/articles/PMC137435/
Our experiment showed that 5U and 10U of Cre recombinase produced a reduction of linear DNA (Kanamycin resistance gene flanked with loxP and lox66) when compare with the control DNA (No Cre recombinase) and 1U of Cre recombinase, as is showed in the figure A. It was also showed an increase in the plasmid form DNA (upper band at 2kb), as is showed in figure B.
The conclusion was that we can use this loxP-lox66 mechanism in our design and we will need at least 5U of Cre recombinase for any in vitro experiments.
In vivo assay
We proved that we can circularize a fragment of DNA (Kanamycin resistance gene) flanked by a loxP and a lox66 sites in vitro, so we decided to test our machine using
codnkjjjjjjjjjjj
We found one colony growing in a LB plate with kanamycin, showing that the fusion was possible. Which is really a good result, because the pGEM plasmid has no RBS, so it is reported that it can be use to express but the expression is really low.
Based on the report made by the igem2010 UT-Tokyo team and some papers, the lox71 (BBa_I718017) from the registry was wrong, it had a cg instead a gc in its center sequence that is between the arms.
Apparently, this part was corrected by the iGEM11_WITS_CSIR_SA team (BBa_K537020), but the sequence from this group had the same error. The corrected part from iGEM11_Tokyo_Tech (BBa_K649205) was right but no DNA was available in the registry. So we decided to synthesized it and test it, using the proper sequence described by http://www.ncbi.nlm.nih.gov/pmc/articles/PMC137435/, and we submitted the correct DNA to the Registry.
 "
"