Team:LMU-Munich/Data
From 2012.igem.org
| Line 54: | Line 54: | ||
===Bacillus BioBrick Box - Promoters=== | ===Bacillus BioBrick Box - Promoters=== | ||
| - | [[File:Auswertung Anderson promoters.png|thumb|right|400px|Fig. 1: | + | [[File:Auswertung Anderson promoters.png|thumb|right|400px|Fig. 1: Luminescence measurement of Anderson promoters in the reporter vector pSB<sub>''Bs''</sub>3C-''luxABCDE''. OD<sub>''600''</sub> (right), LUMI (center) and OD<sub>''600''</sub> per LUMI (left) depending on the time (h) are shown for two different clones (green/blue). Data derive from three independent experiments. Curves were fitted over each other (t=0, OD<sub>''600''</sub>=0,3) and smoothed by taking average of three neighboring values.]] |
Eleven (J23100,J23101, J23102, J23103, J23106, J23107, J23113, J23114, J23115, J23117, J23118) of the nineteen promoters of the Anderson collection were evaluated in the reporter vector pSB<sub>''Bs''</sub>3C-''luxABCDE'' from the BioBrickBox containing the lux operon as a reporter for promoter activity. The gene expression which correlates to the promoter activity leads to the expression of the lux operon with the luciferase. The luminescence which is produced by the luciferase can be measured with the plate reader (BioTek). | Eleven (J23100,J23101, J23102, J23103, J23106, J23107, J23113, J23114, J23115, J23117, J23118) of the nineteen promoters of the Anderson collection were evaluated in the reporter vector pSB<sub>''Bs''</sub>3C-''luxABCDE'' from the BioBrickBox containing the lux operon as a reporter for promoter activity. The gene expression which correlates to the promoter activity leads to the expression of the lux operon with the luciferase. The luminescence which is produced by the luciferase can be measured with the plate reader (BioTek). | ||
| Line 62: | Line 62: | ||
To measure the activity not only with the lux reporter operon, four promoters were cloned into the reporter vector pSBBs1C-lacZ to do beta-galactosidase assays and then to compare the results of the strength of these promoters in B. subtilis. | To measure the activity not only with the lux reporter operon, four promoters were cloned into the reporter vector pSBBs1C-lacZ to do beta-galactosidase assays and then to compare the results of the strength of these promoters in B. subtilis. | ||
| - | [[File:Auswertung_plate_reader_andere_promotoren.png|thumb|right|400px|Fig. 2: | + | [[File:Auswertung_plate_reader_andere_promotoren.png|thumb|right|400px|Fig. 2: Luminescence measurement of the constitutive ''Bacillus'' promoters P<sub>''liaG''</sub> and P<sub>''lepA''</sub> in the reporter vector pSB<sub>''Bs''</sub>3C-''luxABCDE''. OD<sub>''600''</sub> (right), LUMI (center) and LUMI per OD<sub>''600''</sub> (left) depending on the time (h) are shown for two different clones (green/blue). Data come from three independent experiments. Curves were fitted over each other (t=0, OD<sub>''600''</sub>=0,3) and smoothed by taking average of three neighboring values.]] |
The constitutive promoters P<sub>''liaG''</sub> and P<sub>''lepA''</sub> were evaluated in the reporter vector pSB<sub>Bs</sub>3C-<i>luxABCDE</i> which contains the ''lux'' operon. | The constitutive promoters P<sub>''liaG''</sub> and P<sub>''lepA''</sub> were evaluated in the reporter vector pSB<sub>Bs</sub>3C-<i>luxABCDE</i> which contains the ''lux'' operon. | ||
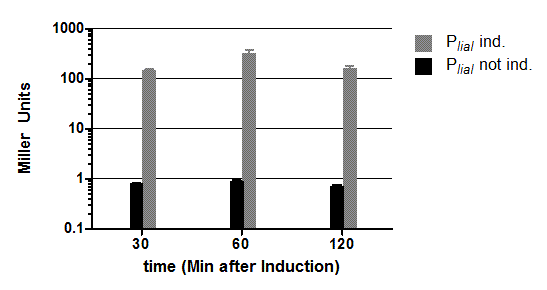
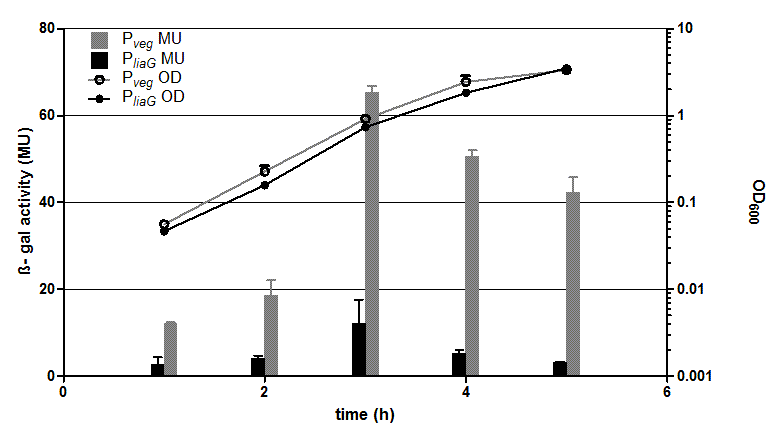
| - | [[File:Englisch_Auswertung_PliaG_Pveg.png|thumb|right|400px|Fig. 3: β-galactosidase assay and growth curve P<sub>''liaG''</sub> (black) and P<sub>''veg''</sub> (grey) in the reporter vector pSB<sub>''Bs''</sub>1C-''lacZ''. β-galactosidase activity (Miller Units)and growth curve are the average of two independant clones. Experiment shows representative data which was | + | [[File:Englisch_Auswertung_PliaG_Pveg.png|thumb|right|400px|Fig. 3: β-galactosidase assay and growth curve P<sub>''liaG''</sub> (black) and P<sub>''veg''</sub> (grey) in the reporter vector pSB<sub>''Bs''</sub>1C-''lacZ''. β-galactosidase activity (Miller Units)and growth curve are the average of two independant clones. Experiment shows representative data which was obtained in the same way from three independent experiments.]] |
| Line 72: | Line 72: | ||
[[File:Englisch Auswertung PliaI.png|thumb|right|400px|Fig. 4: ]] | [[File:Englisch Auswertung PliaI.png|thumb|right|400px|Fig. 4: ]] | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| Line 185: | Line 171: | ||
We induced our germination-mutant strains to sporulate in Difco sporulation media. Then we measured the germination rate of mutant spores in a germination assay. | We induced our germination-mutant strains to sporulate in Difco sporulation media. Then we measured the germination rate of mutant spores in a germination assay. | ||
| - | [[File:germination_assay_plates_720_.jpg|frame|Fig. 5: Comparison of colony growth on LB-agar (plus antibiotics) plates. Upper plates are plated with DSM cell/spore cultures that have been heated at 80°C for 1 hour to kill living cells (but does not affect spores). This prevents cells from forming colonies. Therefore, all colonies observed are from germinated spores. Lower plates contain cultures which have not been heated. Growth on these plates is from either cells or spores, and demonstrates that strains are able to grow. WT168 has no germination knockouts; strains B40-B43 have triple knockouts; strains B46 and B47 have quadruple knockouts; ''Spo0A'' has a knockout of the sporulation gene ''Spo0A'' (replaced with a tet cassette). WT168 is a positive control; ''Spo0A'' is a negative control. <br /> Strains: <br /> '''B40''' -- ''cwlD''::kan, ''sleB''::mla, ''cwlJ''::spec <br /> '''B41''' -- ''cwlD''::kan, ''sleB''::mls, ''gerD''::cat <br /> '''B42''' -- ''cwlD''::kan, ''cwlJ''::spec, ''gerD''::cat <br /> '''B43''' -- ''gerD''::cat, ''sleB''::mls, ''cwlJ''::spec <br /> '''B46''' -- ''cwlD''::kan, ''cwlJ''::spec, ''gerD''::cat, ''sleB''::mls <br /> '''B47''' -- ''gerD''::cat, ''sleb''::mls, ''cwlJ''::spec, ''cwlD''::kan]] | + | [[File:germination_assay_plates_720_.jpg|frame|center|Fig. 5: Comparison of colony growth on LB-agar (plus antibiotics) plates. Upper plates are plated with DSM cell/spore cultures that have been heated at 80°C for 1 hour to kill living cells (but does not affect spores). This prevents cells from forming colonies. Therefore, all colonies observed are from germinated spores. Lower plates contain cultures which have not been heated. Growth on these plates is from either cells or spores, and demonstrates that strains are able to grow. WT168 has no germination knockouts; strains B40-B43 have triple knockouts; strains B46 and B47 have quadruple knockouts; ''Spo0A'' has a knockout of the sporulation gene ''Spo0A'' (replaced with a tet cassette). WT168 is a positive control; ''Spo0A'' is a negative control. <br /> Strains: <br /> '''B40''' -- ''cwlD''::kan, ''sleB''::mla, ''cwlJ''::spec <br /> '''B41''' -- ''cwlD''::kan, ''sleB''::mls, ''gerD''::cat <br /> '''B42''' -- ''cwlD''::kan, ''cwlJ''::spec, ''gerD''::cat <br /> '''B43''' -- ''gerD''::cat, ''sleB''::mls, ''cwlJ''::spec <br /> '''B46''' -- ''cwlD''::kan, ''cwlJ''::spec, ''gerD''::cat, ''sleB''::mls <br /> '''B47''' -- ''gerD''::cat, ''sleb''::mls, ''cwlJ''::spec, ''cwlD''::kan]] |
The plate growth demonstrates the inability of our mutant spores to germinate. We can say that fewer than 1 out of 3x10^7 spores of strains B40, B41, B43, B46, and B47 germinated. | The plate growth demonstrates the inability of our mutant spores to germinate. We can say that fewer than 1 out of 3x10^7 spores of strains B40, B41, B43, B46, and B47 germinated. | ||
{{:Team:LMU-Munich/Templates/Page Footer}} | {{:Team:LMU-Munich/Templates/Page Footer}} | ||
Revision as of 13:51, 11 September 2012
The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

Data
Here you will find all of our project data. For our big breakthroughs, or to follow specific projects, see the individual project pages:
| +--+--+--+--+--+--+-- | +--+--+--+--+-- | +--+--+--+--+--+-- | +--+--+--+--+--+-- |

Bacillus BioBrickBOX |

SporeCoat FusionProteins |

Germination STOP |
Bacillus BioBrick Box - Promoters
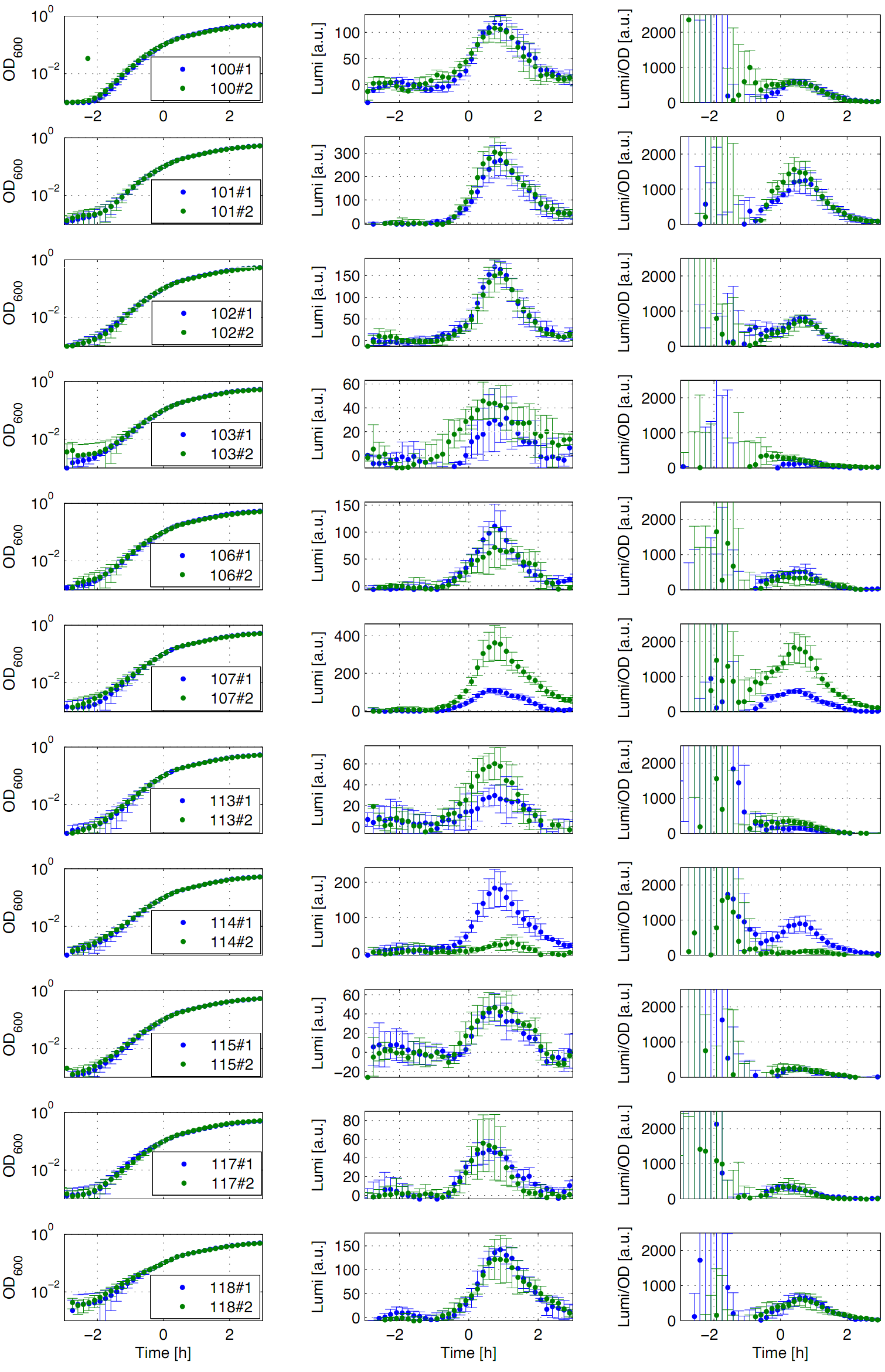
Eleven (J23100,J23101, J23102, J23103, J23106, J23107, J23113, J23114, J23115, J23117, J23118) of the nineteen promoters of the Anderson collection were evaluated in the reporter vector pSBBs3C-luxABCDE from the BioBrickBox containing the lux operon as a reporter for promoter activity. The gene expression which correlates to the promoter activity leads to the expression of the lux operon with the luciferase. The luminescence which is produced by the luciferase can be measured with the plate reader (BioTek).
To measure the activity not only with the lux reporter operon, four promoters were cloned into the reporter vector pSBBs1C-lacZ to do beta-galactosidase assays and then to compare the results of the strength of these promoters in B. subtilis.
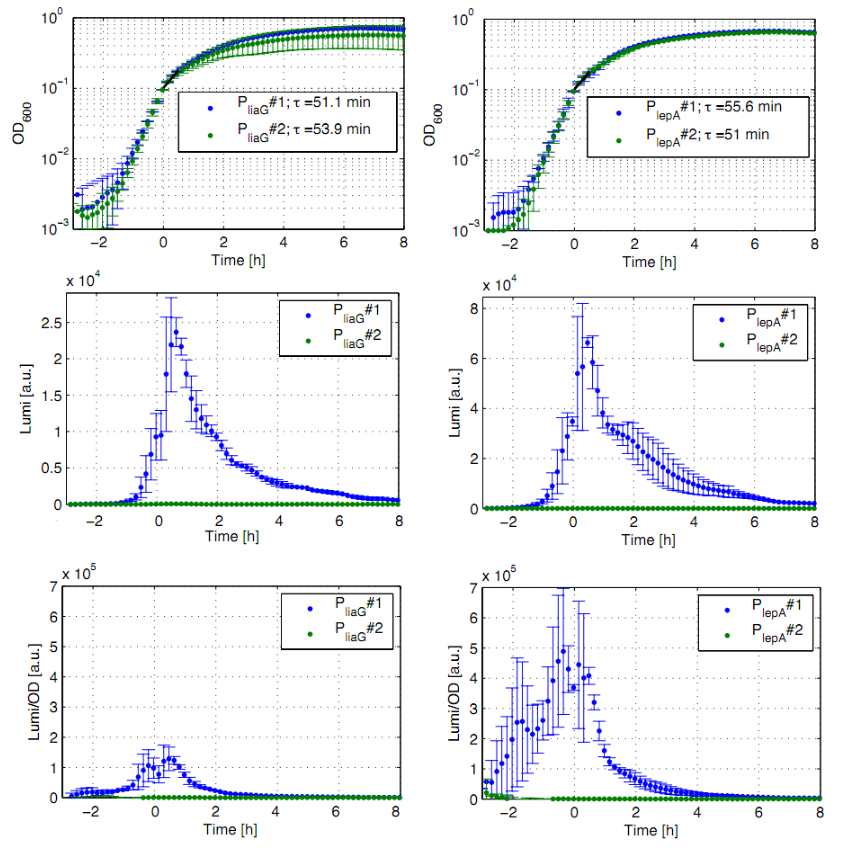
The constitutive promoters PliaG and PlepA were evaluated in the reporter vector pSBBs3C-luxABCDE which contains the lux operon.
Germination Stop
We induced our germination-mutant strains to sporulate in Difco sporulation media. Then we measured the germination rate of mutant spores in a germination assay.

Strains:
B40 -- cwlD::kan, sleB::mla, cwlJ::spec
B41 -- cwlD::kan, sleB::mls, gerD::cat
B42 -- cwlD::kan, cwlJ::spec, gerD::cat
B43 -- gerD::cat, sleB::mls, cwlJ::spec
B46 -- cwlD::kan, cwlJ::spec, gerD::cat, sleB::mls
B47 -- gerD::cat, sleb::mls, cwlJ::spec, cwlD::kan
The plate growth demonstrates the inability of our mutant spores to germinate. We can say that fewer than 1 out of 3x10^7 spores of strains B40, B41, B43, B46, and B47 germinated.
 "
"