Team:Washington/Optogenetics
From 2012.igem.org
| Line 90: | Line 90: | ||
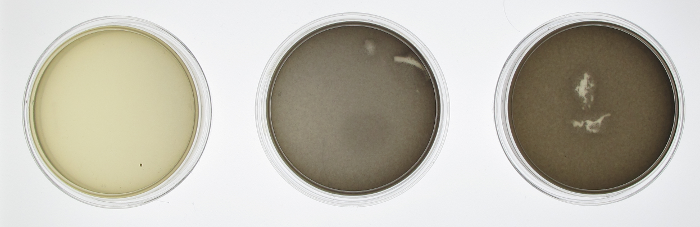
[[File:20120927 big dots cropped experimentwells resized.png|border|400px|left|thumb|Plate Experiment: Columns 1-5: constant exposure to green light. Columns 6-7: flashing red light. Columns 8-12: constant exposure to green light. Rows are replicates.]] | [[File:20120927 big dots cropped experimentwells resized.png|border|400px|left|thumb|Plate Experiment: Columns 1-5: constant exposure to green light. Columns 6-7: flashing red light. Columns 8-12: constant exposure to green light. Rows are replicates.]] | ||
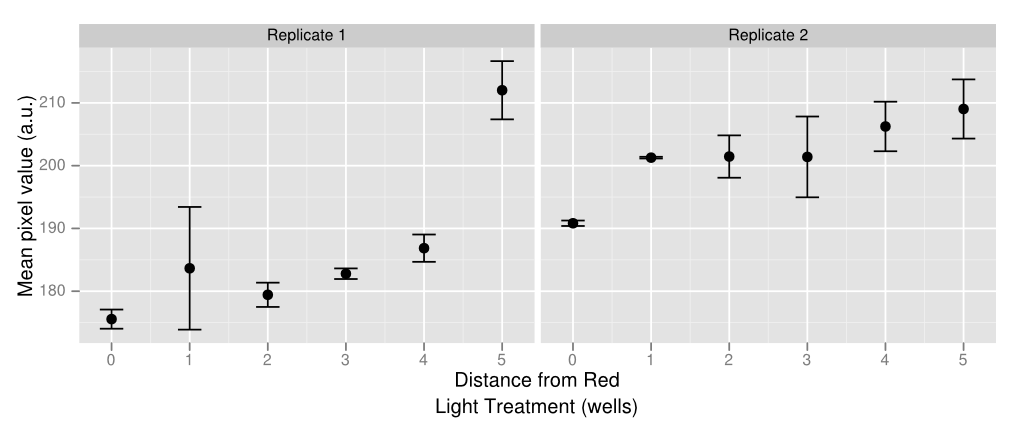
[[File:20120927 big dots cropped experimentwells firstroi ddply plot.png|border|500px|right|thumb|Image analysis of the plate experiment. The y axis is well darkness and the x axis is distance from the center of the plate. 0 is columns 6 and 7, 1 is 5 and 8, and so on. Wells closest to the center (red light, reduces lacZ expression) are lighter than wells that are farther away, indicating a smaller amount of cut S-Gal.]] | [[File:20120927 big dots cropped experimentwells firstroi ddply plot.png|border|500px|right|thumb|Image analysis of the plate experiment. The y axis is well darkness and the x axis is distance from the center of the plate. 0 is columns 6 and 7, 1 is 5 and 8, and so on. Wells closest to the center (red light, reduces lacZ expression) are lighter than wells that are farther away, indicating a smaller amount of cut S-Gal.]] | ||
| - | + | <br> | |
| - | + | <br> | |
| - | + | <br> | |
| - | + | <br> | |
| - | + | <br> | |
| - | + | <br> | |
| - | + | <br> | |
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
---- | ---- | ||
<h1 id='Future'>Future Directions<html><a href="#Background"><font size="3">[Top]</font></a></html></h1> | <h1 id='Future'>Future Directions<html><a href="#Background"><font size="3">[Top]</font></a></html></h1> | ||
Revision as of 04:11, 3 October 2012
Optogenetics: Shedding light on a hands-free approach to protein regulation
Background
Bacterial metabolic pathways constitute a foundation on which to build biological processes that perform useful tasks such as the production of drugs, biofuels, or the degradation of harmful compounds. Often, to achieve an optimally tuned system, expression levels of each component must be varied over a wide range. The burgeoning field of optogenetics affords researchers the ability to control gene expression with light. In addition to being low cost, controlling of gene expression with light has a number of advantages over standard chemical methods of gene control. Among these are the ability to finely tune induction levels through changes in intensity, as well as the ability to quickly and completely remove the input. Further, many light-induced expression systems are reversible depending on the wavelength used for illumination. Thus one of the goals of the 2012 iGEM team is to implement a light inducible system which we hoped to apply to the tuning of multiple metabolic pathways. In addition, we wanted to make the tools to control optogenetic systems readily available and easier to access.
Methods[Top]
Characterization of a light inducible protein expression system for the tuning of biological pathways
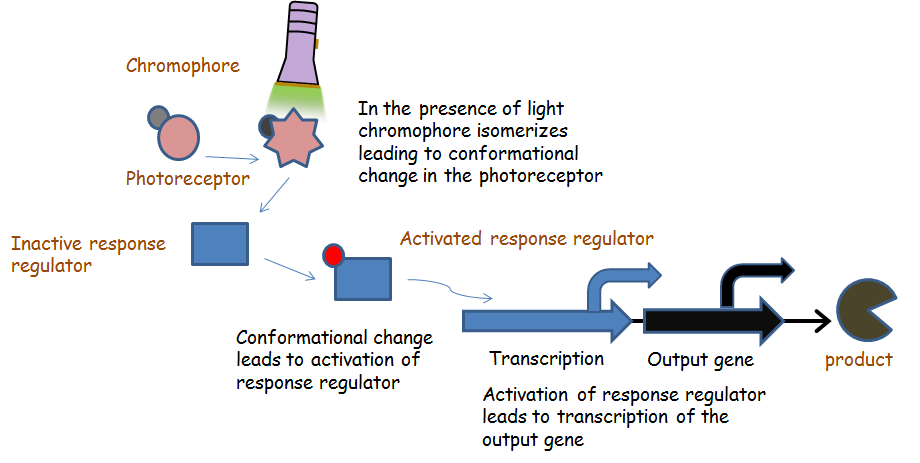
Chromatic tuning is based off light-induced protein interactions. The most basic of these systems is a monochromatic system, in which a specific wavelength of light changes a protein's conformation temporarily, causing it to interact differently with its environment. Our system's protein was a fusion the genes CcaR and CcaS with a chromophore, and changed conformation in response to green light (λ=535nm), and red light (λ=672nm). When stimulated by green light, the protein exhibits improved autophosphorylation, and the phosphorylated protein binds to promoter region PcpcG2 and activates the downstream gene. Red light, on the other hand, changes the protein conformation such that it resists phosphorylation, thus not binding to the promoter and not allowing gene expression. Any gene placed after the promoter can be theoretically controlled by the exposure of λ=535nm and λ=637nm light to the system.
Unfortunately, light-induced protein interactions require a lot of work to characterize. In the past, experiments have involved elaborate set-ups like light masks, or specific-wavelength lasers. When beginning our experiment, we set out to engineer a system that would produce light both modular in position and in color. The result was a tablet app, e. colitune(light), described in detail in the following section.
Building Optogenetic Tools
The current swath of tools available to illuminate bacterial cultures in a controlled manner often necessitate the purchase of specialized equipment or materials. Further, devices must often be both constructed, and fully characterized by the researcher prior to conducting any experiments to ensure reproducibility. To circumvent these problems, we sought to develop a tablet based application for the Android platform which affords a fully customizable method for illumination of bacterial plate cultures inexpensively and reproducibly. In recent years, tablet computers have become increasingly ubiquitous. Each year sees the release of many new tablets, including some that are relatively cheap. Further, many tablets operate on the Android platform, allowing us to develop a freely available app with many features we believe will be useful for future optogenetics studies.
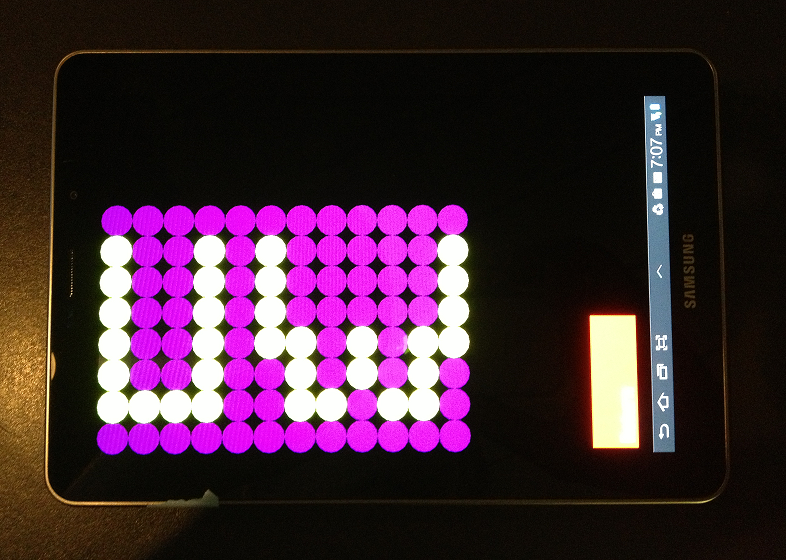
Tools currently available for testing of optogenetic systems, that is one that can shine different wavelengths of light of different well sizes at different frequencies are currently very costly and difficult to obtain. We wanted to make optogenetics more accessible to the scientific community and to do that we decided to create a tool that would make testing these optogenetic systems much more cost effective and easier to obtain. To test our bacteria under various colours of lighting we created the android tablet app. The application can generate different wavelengths of light (currently blue, green and red) in shapes of wells or plates and with varying frequencies and wait times between colors. To use the application we simply chose well or plates, and then chose wait time between flashes of colors in milliseconds of wait time and color intensities of each color from 0-100 (max needs to explain this and what each of these numbers mean.) The bacteria can then be grown overnight placed on top of the app. The app is currently available in the google play store for free and provides a convenient way for future iGEM teams and scientists to test their optogenetic systems. Because iGEM teams and labs will have a set budget, we hope that this tool becomes useful to all that want to pursue science.
E.colight app feature list:
- Petri Dish / 96 Well Plate Formats available
- Red, Blue, and Green light available
- Intensity of light delivered can be modulated by:
- Frequency of Flashing
- Changing light intensity
- Well sizes can be changed (in 96 well plate format only)
Experimental Description
For the bacteria density gradient experiment, we grew an overnight culture of JT2 cells transformed with pJT122 and pJT106b overnight in buffered LB. Cultures were grown in tubes wrapped with aluminum foil to ensure no activation by light occurred during growth. We then diluted the cultures in a 1:5 serial dilution to a final dilution of X. After planted on the 96-well plate in a row, the bacteria were shined continuously by tablet green light mode. The experiment was carried in 37 C incubator and the period is 15-hour long, which we concluded from long-time repeated tests. The result showed obvious gradient on the darkness of spots in the wells and illustrated the bacteria gradient can control the pigment produced.
From the Tabor paper, we found red light could inhibit the pigment production process. So we followed the ~2-inch petri dish mixture protocol indicated in web page and set control experiment up. We set one plate in green light, one in red light, and one covered with aluminium foil, all of which placed in 37 C incubator. After 15-hour wait and quantification, the result showed the lowest intensity in red light and higher one in green light. Then the red light inhibition system in this JT2 bacteria is confirmed.
During the control test on one 96-well plate between green light and red light comparison, we found the certain green project area was lighter than some wells far away from the red light. After several repeated tests, we were sure that this was not a coincident, but a tablet light leak. Since the light projected from tablet was not vertically on each well and the 96-well plate had no smooth surface, the light would reflect and spread among wells, which lead to light leak. In general, two solutions were come up: one is to use smaller area of light in each well, which is one of the function you can see in the app now; the other is to use a rubber gasket between plate and tablet to decrease light amount and reflection. The tests confirmed the rubber gasket is a good way to solve the light leak by tablet.
Three typical experiments are all stated in Result section.
Results Summary [Top]
The finished app was capable of flashing series of blue, green, or red light, or none at all, for an amount of time up to [maximum wait time]. Despite originally intending to compensate for light intensity with this, we learned that programming intensity of light was easy, so the app comes with both for easier tuning.
The app is currently able to use two culture formats. One is a 96-well plate, which contains 96 separately-controllable light sources. The other is a standard petri dish, which acts as a single light source. During testing, we found that light had a tendency to leak between wells of a 96-well plate despite the spacing between wells and the LEDs on the app. To somewhat compensate for this, an option for well size was added that allows the wells to be adjusted between 1 and 100% of their normal size. This limits leakiness between wells while still allowing for strong induction.
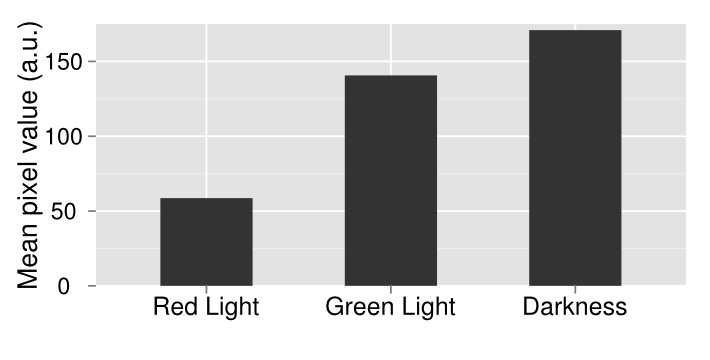
We wanted to characterize the multichromatic system put together by Tabor et. al. In order to confirm the functionality of our light sensor we did a series of experiments. Our first experiment involved simply plating our bacteria onto three plates and then growing them under illumination of different colors of light or no light at all in the case of the control. We see that there is a clear difference between the plates. On the left, when the plate was grown on red light, there was complete repression of the reporter gene, therefore a white plate. In the middle, the plate was grown under illumination of green light and showed increased gene expression afterwards. The control plate which was grown in aluminum foil shows high levels of gene expression; higher than what should be expected of under normal conditions. There should be intermediary amounts of transcription, somewhere between the gene being turned on and being off. However, due to a leaky promoter transcription occurs via the response regulator more often than is expected.
In order to make a better comparison between the three different plates, they were quantified through the use of imageJ. The average pixel density was taken for each plate and then graphed. The quantified data gives a more careful look at our results that are not instantly apparent by visually just looking at it. By looking at the quantified data it’s apparent that grown in the dark, there appears to be more gene expression. What should be expected of this system is that there should be the least amount of gene expression under red light, which was accomplished, mediary amounts for the control plate and maximum gene expression when grown under red light. Due to our results, we have concluded that the system works well under red light repression, since red light is able to almost completely halt gene expression.
To create some kind of gradient for the multichromatic system, we decided to use a 96 well plate and the app. There are rows with constant green light excitation, two rows in the middle that flash and end rows that have constant green light on. The plate is darkest on either end with a gradient that lightens up as it gets closer to the middle, where red and green light flash, inducing intermediary amounts of gene expression. Ideally there should only be lighter colors of wells in columns 6 and 7. However, there appears to be some leakiness with the utilization of the light app as some of this light has managed to leak to other wells, which creates a sort of gradient with darkest wells on either side and becoming lighter towards the middle. Pipetting error also affects the data as some of the wells have more colonies than others which leads to higher pixel values even if gene expression may not necessarily be higher. Using soft agar can help resolve this problem in the future. The quantified data above was made by using ImageJ and measuring average pixel value of each plate. Generally there is a gradient of increased pixel value as the well is further away from the red light. Both replicates show similar results, even though replicate one has slightly ,more dramatic increases while replicate one appears to have a steadier rise. Because of leakiness, some of the flashing red light that was supposed to be confined to wells 6 and 7 have leaked over to nearby wells, causing some gene repression even when there shouldn’t necessarily have been any. However the results show that gene repression is extremely sensitive to red light and a gradient can be created from various levels of red light relatively easily. That being said, gene control isn’t completely “on” or “off” but also holds the benefit of having various levels of “on” and “off”.
Future Directions[Top]
We want to work more with Tabor’s multichromatic system, improving the light sensor available so that we can biobrick them later and have those pieces readily available for use. We suspect that a big problem with the light sensor right now is the leaky promoter, which leads to more transcription than we would normally expect in dark conditions. The promoter should be modified accordingly, either by fixing any present mutation or choosing to use a new promoter entirely for various reasons. Also to improve characterization of the sensor itself, in the future soft agar should be used which will allow more sufficient spread of bacteria, which should minimize differences in bacteria colonies in each well, allowing better data collection with imageJ. There also appears to be some leakiness between wells that should be addressed more appropriately in the future. A suggestion was made to use a rubber stopper with holes in it on top of the app to try to minimize leakiness of light between different wells.We also want to work with different light sensors to improve functioning and to use light sensors to promote various gene expression of different proteins.
The app is readily available and can be downloaded onto any android OS system through the Google play store. Currently we have access to the tablet system with the app on it so we will want to use this app to further test our bacteria in the future to validate for functionality and to control genetic expression.
Sources [Top]
Levskaya, A. et al. Synthetic biology: Engineering Escherichia coli to see light. Nature 438, 441–442 (2005).
Tabor, J. J., Levskaya, A. & Voigt, C. a Multichromatic control of gene expression in Escherichia coli. Journal of molecular biology 405, 315–24 (2011).
 "
"