Team:Washington/Flu
From 2012.igem.org
| Line 92: | Line 92: | ||
== Yeast Surface Display == | == Yeast Surface Display == | ||
| - | To test our designs, we used yeast surface display, which allowed us to analyze a subset of our constructs at 4 different antigen (HA) concentrations: 0,1, 10, and 100 nM. In this process, our mutant plasmids were first expressed in yeast and grown in SDCAA media. Next, | + | To test our designs, we used yeast surface display, which allowed us to analyze a subset of our constructs at 4 different antigen (HA) concentrations: 0,1, 10, and 100 nM. In this process, our mutant plasmids were first expressed in yeast and grown in SDCAA growth media. Next, cells were grown in SCGAA media which contained glucose and galactose, required for the activation of the galactose inducible promoter on the yeast plasmid. This allows the mutant plasmid to be expressed on the yeast surface. All yeast cell samples were standardized to an OD600 of 2, and then labeled using different amounts of a single type of biotinylated antigen (HA). After an incubation step, the samples were then washed to remove unbound antigen. The samples were then labeled with Streptavidin-PE (to monitor binding of antigen, see in red in the diagram below) and anti-cymc FITC (to monitor surface expression, seen in green in the diagram to the right). |
| - | [[Image: | + | [[Image:ysd.png|border|300px|right|thumb|Yeast surface display]] |
| - | + | <br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
We then analyzed the fluorescent signature of the yeast cells using flow cytometery. This generates graphs of the mean PE fluorescence of the surface displayed population of cells as a function of antigen, and the KD is calculated that fits to that data assuming 1:1 binding stoichiometry. | We then analyzed the fluorescent signature of the yeast cells using flow cytometery. This generates graphs of the mean PE fluorescence of the surface displayed population of cells as a function of antigen, and the KD is calculated that fits to that data assuming 1:1 binding stoichiometry. | ||
Revision as of 22:52, 30 September 2012
Background
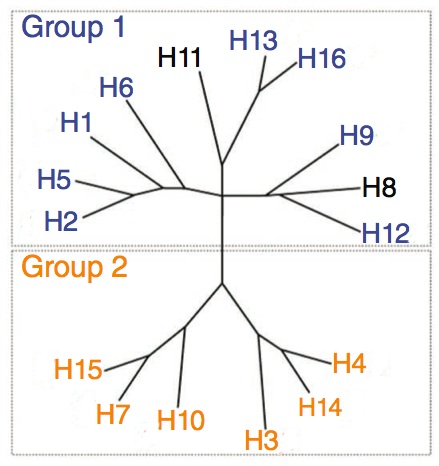
The influenza virus, also known as the flu, is a deadly virus that has decimated large populations of various species including humans. However, even with the advent of vaccinations, high rates of mortality still occur because of the constant evolution of the virus. Variations within each strain of influenza are found within the viral surface proteins that coat their surfaces. One such protein is Hemagglutinin (HA), which is represented by the H when describing flu subtypes (H1N1, H2N3, etc). Hemagglutinin is primarily involved in viral transfer as it mediates the binding and entry into a host cell through a sialic acid linkage. Each strain of Influenza is characterized by it's variant of Hemagglutinin. There are 16 known subtypes of HA; these subtypes give rise to the vast multitude of strains that exist to today, as well as future emergent strains. These subtypes usually only differ by a relatively few number of mutations. The World Health Organization uses the history of influenza breakouts to predict upcoming strains which could threaten to cause future pandemics and vaccinations are made using these predictions.
There are 16 subtypes of HA. They are divided into two groups based on their phylogenicity. However, our team only work with the HAs in group 1.
In 2011, Fleishman et al., detailed two small proteins which had binding capabilities to various subtypes of hemagglutinin. In 2012, Whitehead et al., optimized the flu binders and showed that HB80.4 was broadly binding, meaning it bound to multiple HAs with high affinity. They also showed that HB36.5 bound preferentially to HA 1.
We aimed to further optimize the binding of HB80.4 to the group 1 HAs, but knowing that HB80.4 was already broadly binding to many HAs, we wanted to focus our efforts on the flu binder HB36.5 which until had not yet been tested against any other HAs is group 1.
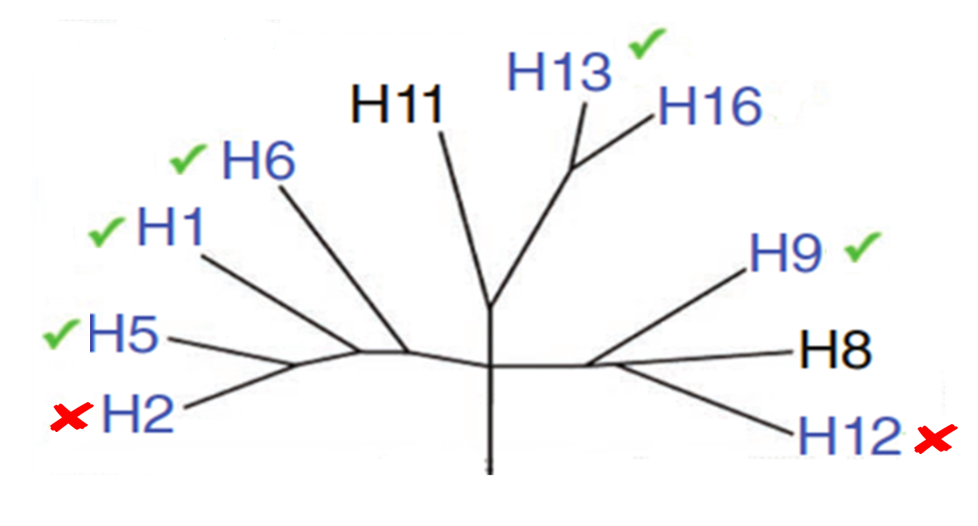
Therefore, we wanted to test HB36.5 against the other group 1 hemaglutinins and asses its native binding capabilities. After testing, we discovered that in addition to hemagglutinin 1, HB36.5 bound to H5, H5, H9 and H13. Binding was observed to hemagglutin 2 or hemaglutin 12. We then proceeded create subtype specific flu binders for these two HAs. Having two broadly – binding flu binders, with subtype specific capabilities would allow for enhanced rapid and reliable global tracking of current and future Influenza strains.
Methods [Top]
Method of computational design
Gathering information about the structures of Hemagglutinin
Since we wanted to improve the binding of HB80.4 and HB36.5 to specific HAs, we had to understand the structural differences of the many subtypes of hemagglutinin. Initially, we searched for specific hemagglutinin structures on the Protein Data Bank (http://www.rcsb.org/pdb/home/home.do). If we found the desired hemagglutinin structures, we downloaded the associated PDB file and made a model of the binder-HA complex in Pymol. These model would then later be used in Foldit.
If we could not find the exact structure of a particular hemagglutinin, we then searched for the protein sequence of that hemagglutinin on the National Center for Biotechnology Information (www.ncbi.nih.gov). We then aligned the protein sequence of the structurally unknown hemagglutinin we obtained to that of the H1 HA (which has a known structure) by using Clustalw2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Next, we created homology models in FoldIt of the desired hemaggluttinin by changing the side chains of the H1 HA in Foldit according to the protein sequence differences we found.
Making model of Foldit
Crystal stuctures of HB36.3 and HB80.4 in complex with H1HA are available on the PDB as PDB ID 3R2X and 4EEF, respectively. To create models our the binder bound to different HAs, we aligned our alternative HA structures to the H1HA in the published crystal structures using PYMOL. Then, we would record the difference in side chains between that specific hemagglutinin and H1HA and use FoldIt to implement those changes on the known crystal structures. The models we created are linked below:
Proposing mutation on the model
After preparing the Foldit model, we proposed mutations on the original flu binder in order to improve its binding to the desired hemagglutinin.
There is a score total in Foldit which indicates the energy of the whole protein structure. It is our understanding that the protein structure is more stable when its energy is lower. Thus, our primary goal was to decrease the score total in Foldit. We did that in three different ways, filling the holes, making space, and balancing the electrostatic. Holes that did not exist in H1HA would be created with the replacement of different side chains. We would mutate the corresponding side chain on the flu binder so that it extends out and fill a hole on the hemagglutinin. Making space is the exact opposite of filling the holes. Some side chains of certain hemaggluttinins would be more projected out in space than that of hemagluttinin one. Therefore, we would mutate the corresponding side chains of the flu binder in order to provide room for the more extended side chains. Some variations of side chain on hemaggluttinin causes some electrostatic changes. We would mutate the corresponding side chains of the flu binder so as to counter balance the electrostatic changes. For example, if a certain area on the helix is changed to be more electronegative, we would introduce an electropositive side chain on the flu binder for improving the binding and vice versa.
| File:HB80 H12 model.zip |
|---|
| File:HB80 H9 model.zip |
|---|
| File:HB80 H5 model.zip |
|---|
| File:HB36 H13 model.zip |
|---|
| File:HB36 H12 model.zip |
|---|
| File:HB36 H9 model.zip |
|---|
| File:HB36 H6 model.zip |
|---|
| File:HB36 H5 model.zip |
|---|
| File:HB36 H2 model.zip |
|---|
Kunkel Mutagenesis
The first step to producing our specially designed flu binders was to change the original flu binding protein with our desired amino acid substitutions.
We designed mutagenic oligonucleotide primers that would anneal to the original flu binding protein and incorporate point mutations that, when expressed, would result a variant of the binder with the desired amino acid shift.
To incorporate these mutations, we first isolated single stranded DNA (ssDNA) of our vector harboring the original binding sequence. To do this we infected cells with bacteriophage M13, which packages its own ssDNA genome identified by length, and so in tandem packaged our vector in single stranded form. We then harvested the phage from the lysed culture of E. coli, and extracted our single stranded vector DNA.
Next, we annealed and extended our mutagenic oligos to incorporate the specified mutations into the newly synthesized antisense strand. This hybrid vector was transformed into E. coli that degraded the original uracil-containing DNA and replaced it with sections complementary to the mutagenized strand.
Yeast Surface Display
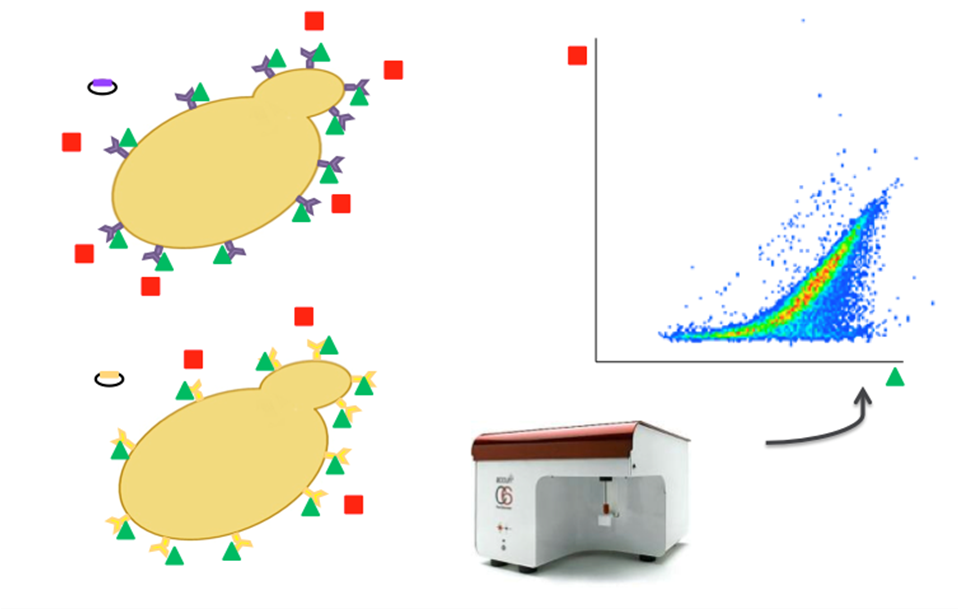
To test our designs, we used yeast surface display, which allowed us to analyze a subset of our constructs at 4 different antigen (HA) concentrations: 0,1, 10, and 100 nM. In this process, our mutant plasmids were first expressed in yeast and grown in SDCAA growth media. Next, cells were grown in SCGAA media which contained glucose and galactose, required for the activation of the galactose inducible promoter on the yeast plasmid. This allows the mutant plasmid to be expressed on the yeast surface. All yeast cell samples were standardized to an OD600 of 2, and then labeled using different amounts of a single type of biotinylated antigen (HA). After an incubation step, the samples were then washed to remove unbound antigen. The samples were then labeled with Streptavidin-PE (to monitor binding of antigen, see in red in the diagram below) and anti-cymc FITC (to monitor surface expression, seen in green in the diagram to the right).
We then analyzed the fluorescent signature of the yeast cells using flow cytometery. This generates graphs of the mean PE fluorescence of the surface displayed population of cells as a function of antigen, and the KD is calculated that fits to that data assuming 1:1 binding stoichiometry.
Results Summary [Top]
HB36.5_F317H was predicted to increase HA5 binding but it decreased the binding to HA9. Nevertheless, it can be used for specific hemagglutinin subtypes binder.
HB36.5_H312A was predicted to increase HA2 binding and it did improve binding to HA2.
HB80.4_A280Vwas predicted to decrease HA2 binding but it improved the binding to HA2.
HB80.4_K298E was predicted to increase HA2, HA5, HA6 and HA12 binding and it did improve binding to HA2.
HB80.4_S276H was predicted to increase HA6 binding but it decreased the binding to HA6.
HB80.4_K298S was predicted to increase HA12 binding but it decreased the binding to HA6.
HB80.4_S286K was predicted to increase HA12 binding and it increased the binding to HA12. However, it decreased the binding to HA6.
Future Directions [Top]
<p> We intend to combine the HB36.5 single- point mutants H312A and W313A ,both successfully having increased binding to HA2, to create a superior combinatorial mutant with multiple-fold improvement. Simultaneously, we intend to create a mutant of HB36.5 that will bind H12 preferentially, as HB36.5 does not currently do so.
Parts Submitted [Top]
HB80.4 (Wild-type Flu binder)
HB80.4_S276H
• Predicted to increase HA6 binding.
HB80.4_K298S
• Predicted to increase HA12 binding.
HB80.4_S286K
• Predicted to increase HA12 binding.
HB80.4_K304S
• Predicted to increase HA6 binding.
• Predicted to increase HA12 binding.
• Predicted to increase HA13 binding.
HB80.4_K274R
• Predicted to increase HA13 binding.
HB80.4_A280V
• Predicted to decrease HA2 binding.
HB80.4_K298E
• Predicted to increase HA2 binding.
• Predicted to increase HA5 binding.
• Predicted to increase HA6 binding.
• Predicted to increase HA12 binding.
HB80.4_ K298Q
• Predicted to increase HA13 binding.
HB80.4_Q301K
• Predicted to increase HA2 binding.
HB80.4_Q301R
• Predicted to increase HA2 binding.
• Predicted to increase HA5 binding.
• Predicted to increase HA9 binding.
HB36.5 (Wild-type Flu binder)
HB36.5_A326E
• Predicted to increase HA2 binding.
• Predicted to increase HA5 binding.
• Predicted to increase HA6 binding.
• Predicted to increase HA12 binding.
• Predicted to increase HA13 binding.
HB36.5_A326E + P324K
• Predicted to decrease HA2 binding.
• Predicted to increase HA5 binding.
• Predicted to increase HA6 binding.
• Predicted to increase HA13 binding. HB36.5_F317H
• Predicted to increase HA5 binding.
HB36.5_F317S
• Predicted to increase HA2 binding.
• Predicted to increase HA9 binding.
HB36.5_H312A
• Predicted to increase HA2 binding.
HB36.5_W313A
• Predicted to increase HA2 binding.
 "
"