Team:Wageningen UR/Journal/week18
From 2012.igem.org
Lisamsonne (Talk | contribs) |
(→week 18: 27 August - 2 September) |
||
| (21 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:WUR}} | {{Template:WUR}} | ||
| + | |||
| + | = week 18: 27 August - 2 September = | ||
| + | |||
| + | ''' CCMV ''' | ||
| + | |||
| + | ''27th August'' (Hugo) | ||
| + | <br/> | ||
| + | Isolated VLP’s from the dialysed solution using an ultracentrifuge. | ||
| + | <br/> | ||
| + | Colony PCR on iron inducable biobricks. | ||
| + | <br/><br/> | ||
| + | ''28th August'' (Hugo) | ||
| + | Making TEM pictures. | ||
| + | [[File:KeesvanderArkontheEM.PNG|600px|center|thumb|''Figure 1: Kees van der Ark handling the Electron Microscope.'']] | ||
| + | <br/><br/> | ||
| + | Inoculated 10 mL LB with IPTG_CCMV_NEG JM109 cells. | ||
| + | <br/><br/> | ||
| + | ''29th August'' (Hugo) | ||
| + | <br/> | ||
| + | Sent BBA_J3901 for sequencing with forward sequencing primer (BaseClear). | ||
| + | <br/> | ||
| + | Miniprepped the 10 mL of LB with IPTG_CCMV_NEG JM109 cells. | ||
| + | <br/> | ||
| + | Inoculated 10 mL LB with IPTG_CCMV_NEG JM109 cells. | ||
| + | <br/><br/> | ||
| + | ''30th August'' (Hugo) | ||
| + | <br/> | ||
| + | Inoculated 200 mL LB medium with 5 mL of IPTG_CCMV_NEG cell culture. Induced before OD600 was 0.6 and grew for another 4 hrs. Then pelleted the culture, followed by washing steps. Stored the pellet in the freezer overnight. | ||
| + | <br/> | ||
| + | Digested IPTG_CCMV_NEG plasmid + Jeroen’s IPTG_CCMV_Delta26, IPTG_CCMV_Delta26_His, IPTG_CCMV_His + pSB1C3 with EcoRI + PstI. | ||
| + | <br/> | ||
| + | Ran digestion products on an agarose gel and isolated gel slices of the visible bands. | ||
| + | <br/><br/> | ||
| + | ''31st August'' (Hugo) | ||
| + | <br/> | ||
| + | Disrupted the pelleted cells using a french press and prepared two solutions for dialysis. Ran dialysis over the weekend. | ||
| + | <br/><br/> | ||
| + | ''2nd Septembre'' (Hugo) | ||
| + | <br/> | ||
| + | Purified the gel slices isolated on 30-08. | ||
| + | <br/><br/> | ||
| + | '''TuYV''' | ||
| + | <p align="justify"> | ||
| + | This was the a very frustrating week for the TuYV group. | ||
| + | <br><br> | ||
| + | We retried the second step extension PCR twice, but we always got smear on the gel check later. | ||
| + | <br><br> | ||
| + | Colony PCR with TuYV primers to test last week transformation was done twice as well, however it showed positive results on the negative control, we assumed the TuYV primer was contaminated, later we designed an PCR with different combinations of forward and reverse primers we had without adding any template. On the gel check we found positive results, which confirmed that the primers we had were contaminated. | ||
| + | <br><br> | ||
| + | Colony PCR with sequencing primers to test last week transformation was performed twice. It confirmed our TuYV constructs were not inserted after the IPTG induced promoter. | ||
| + | <br><br> | ||
| + | Three samples( TuYV coat protein, coat protein with His-tag, and coat protein with part of readthrough in the chloramphenical backbone), which showed positive results on the colony PCR, were send out for sequencing on Wednesday. Sequencing results were received on Friday. The results revealed that our TuYV constructs were not in the chloramphenical backbone. | ||
| + | <br><br> | ||
| + | After two mouth's work, we have to start over again. | ||
| + | |||
| + | </p> | ||
| + | |||
| + | |||
| + | |||
| + | ---- | ||
'''Hepatitis B''' | '''Hepatitis B''' | ||
| Line 5: | Line 65: | ||
---- | ---- | ||
''27.August'' | ''27.August'' | ||
| - | * Miniprep of HepB+IPTG in | + | * Miniprep of HepB+IPTG in pSB1C3 backbone in DH5α (transformation from 20 August) to check again wheather there is an insert |
-> the miniprep had a very low yield | -> the miniprep had a very low yield | ||
| Line 15: | Line 75: | ||
''29 August'' | ''29 August'' | ||
| - | * Miniprep of HepB PCR product in | + | * Miniprep of HepB PCR product in pSB1C3 backbone from colony PCR 20.Aug and HepB with his-tag (in pSB1C3 backbone) from colony PCR 20Aug to check again if there is an insert (since colony PCR is not working well sometimes) |
| + | -> the DNA concentration of the minipreps was very low! | ||
* PCR check of the minipreps with both HepB primers as well as sequencing primers | * PCR check of the minipreps with both HepB primers as well as sequencing primers | ||
-> some bands (with the right size) are visible for HepB with the his-tag and HepBwt (original HepB core protein) – but also in the same lane a second band (about the size of the backbone) - no conclusion can be made | -> some bands (with the right size) are visible for HepB with the his-tag and HepBwt (original HepB core protein) – but also in the same lane a second band (about the size of the backbone) - no conclusion can be made | ||
| - | |||
''30 August'' | ''30 August'' | ||
* Inoculate sample 20 from 20July to make a new HepB+IPTG promoter biobrick – stock (cells did not grow) | * Inoculate sample 20 from 20July to make a new HepB+IPTG promoter biobrick – stock (cells did not grow) | ||
| + | -> did not grow | ||
| + | |||
| + | |||
| + | ''31 August'' | ||
| + | * again miniprep of HepB PCR product in pSB1C3 backbone from colony PCR 20.Aug and HepB with his-tag | ||
| + | -> this time the yield was higher | ||
| + | |||
'''Hepatitis B outside modification''' | '''Hepatitis B outside modification''' | ||
| Line 33: | Line 100: | ||
• Whole plasmid PCR | • Whole plasmid PCR | ||
| - | + | [[File:WholePlasmid.JPG|500px|center|thumb|<p align="justify">''Figure 2: Schematic overview of whole plasmid pcr'</p>]] | |
| + | Instead of using the general FW and Rev primers, the whole plasmid can be amplified alternatively. This allows a more controlled Tm and only 1 amplicon instead of 2 which have to be combined. The same steps 1-4 are still required. | ||
| - | + | First try with Phusion polymerase did not have any result. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | First try with | + | |
* 29 August (Kees) | * 29 August (Kees) | ||
| Line 47: | Line 111: | ||
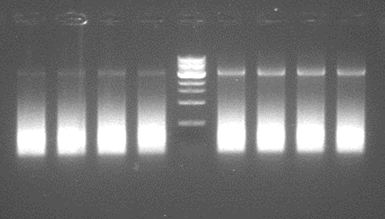
A greadient pcr was performed using Taq polymerase. This can only determine wether the primers do work at a certain, yet for phusion undertermined temperature. Besides, Taq adds an A, causing a mutation in the final step. Therefore, the products can not be used, even if we accept the absence of proofreading in Taq. | A greadient pcr was performed using Taq polymerase. This can only determine wether the primers do work at a certain, yet for phusion undertermined temperature. Besides, Taq adds an A, causing a mutation in the final step. Therefore, the products can not be used, even if we accept the absence of proofreading in Taq. | ||
| + | |||
| + | [[File:GradientPCRstep4.JPG|500px|center|thumb|<p align="justify">''Figure 3: The bands on the gel are as expected somewhat over 3k bp. This proofs this pcr step is possible.'</p>]] | ||
| - | + | ||
• Phusion Polymerase PCR | • Phusion Polymerase PCR | ||
| Line 65: | Line 131: | ||
* 4th (final) step of the PCR reaction with the sample from the gel purification | * 4th (final) step of the PCR reaction with the sample from the gel purification | ||
| - | [[File:4th step PCR hepinsidecoil.png| | + | |
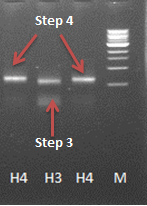
| + | [[File:4th step PCR hepinsidecoil.png|center|200px|thumb|<p align="justify">Figure 4: 3rd and 4th step of the Hep B -inside coil PCR reaction]] | ||
-> this step seemed to work - the gel shows a band at the expected size slighly larger than the band from the 3rd step. | -> this step seemed to work - the gel shows a band at the expected size slighly larger than the band from the 3rd step. | ||
| Line 84: | Line 151: | ||
* 2nd try of the check and amplification of GFP-coil PCR product from 14.Aug | * 2nd try of the check and amplification of GFP-coil PCR product from 14.Aug | ||
-> still no band visible for the GFP check/amplification | -> still no band visible for the GFP check/amplification | ||
| - | * Miniprep of GFP-coil in | + | * Miniprep of GFP-coil in pSB1C3 backbone in JM109 (transformation from 20 August) to check again wheather there is an insert |
| Line 99: | Line 166: | ||
* Transformation of the ligation product with Mach1 (once with the direct ligation product and once with purified ligation product (PCR purification) | * Transformation of the ligation product with Mach1 (once with the direct ligation product and once with purified ligation product (PCR purification) | ||
-> no colonies on the plate | -> no colonies on the plate | ||
| + | |||
| + | |||
| + | ''31 August'' | ||
| + | * again miniprep of GFP-coil in pSB1C3 backbone in JM109 (transformation from 20 August) and HepB with his-tag | ||
| + | -> this time the yield was higher | ||
| + | |||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | '''CCMV Stability experiments''' | ||
| + | |||
| + | ''27th August'' (Mark) | ||
| + | <ul> | ||
| + | <li>pH stability experiment with DLS</li> | ||
| + | <ul> | ||
| + | <li>pH range 4.5 - 7.5</li> | ||
| + | <li>pH change was to sudden not measurable with DLS</li> | ||
| + | </ul> | ||
| + | </ul> | ||
| + | ---- | ||
| + | |||
| + | |||
| + | '''PLRV''' | ||
| + | |||
| + | ---- | ||
| + | Because the expression of PLRV coat proteins was not successful in E. coli DH5α we transformed E. coli BL21 cells with the IPTG+RBS_PLRV_CP constructs (PLRV coat proteins downstream a IPTG inducible promoter plus ribosomal binding site). | ||
| + | After expression, a SDS-PAGE gel was run but again no difference in protein expression was noticed. | ||
| + | |||
| + | |||
| + | [[https://2012.igem.org/Team:Wageningen_UR/Journal/week17 previous week]] [[https://2012.igem.org/Team:Wageningen_UR/Journal/week19 next week]] | ||
Latest revision as of 03:36, 27 September 2012
week 18: 27 August - 2 September
CCMV
27th August (Hugo)
Isolated VLP’s from the dialysed solution using an ultracentrifuge.
Colony PCR on iron inducable biobricks.
28th August (Hugo)
Making TEM pictures.
Inoculated 10 mL LB with IPTG_CCMV_NEG JM109 cells.
29th August (Hugo)
Sent BBA_J3901 for sequencing with forward sequencing primer (BaseClear).
Miniprepped the 10 mL of LB with IPTG_CCMV_NEG JM109 cells.
Inoculated 10 mL LB with IPTG_CCMV_NEG JM109 cells.
30th August (Hugo)
Inoculated 200 mL LB medium with 5 mL of IPTG_CCMV_NEG cell culture. Induced before OD600 was 0.6 and grew for another 4 hrs. Then pelleted the culture, followed by washing steps. Stored the pellet in the freezer overnight.
Digested IPTG_CCMV_NEG plasmid + Jeroen’s IPTG_CCMV_Delta26, IPTG_CCMV_Delta26_His, IPTG_CCMV_His + pSB1C3 with EcoRI + PstI.
Ran digestion products on an agarose gel and isolated gel slices of the visible bands.
31st August (Hugo)
Disrupted the pelleted cells using a french press and prepared two solutions for dialysis. Ran dialysis over the weekend.
2nd Septembre (Hugo)
Purified the gel slices isolated on 30-08.
TuYV
This was the a very frustrating week for the TuYV group.
We retried the second step extension PCR twice, but we always got smear on the gel check later.
Colony PCR with TuYV primers to test last week transformation was done twice as well, however it showed positive results on the negative control, we assumed the TuYV primer was contaminated, later we designed an PCR with different combinations of forward and reverse primers we had without adding any template. On the gel check we found positive results, which confirmed that the primers we had were contaminated.
Colony PCR with sequencing primers to test last week transformation was performed twice. It confirmed our TuYV constructs were not inserted after the IPTG induced promoter.
Three samples( TuYV coat protein, coat protein with His-tag, and coat protein with part of readthrough in the chloramphenical backbone), which showed positive results on the colony PCR, were send out for sequencing on Wednesday. Sequencing results were received on Friday. The results revealed that our TuYV constructs were not in the chloramphenical backbone.
After two mouth's work, we have to start over again.
Hepatitis B
27.August
- Miniprep of HepB+IPTG in pSB1C3 backbone in DH5α (transformation from 20 August) to check again wheather there is an insert
-> the miniprep had a very low yield
28 August
- EM detection of VLPs
After explanation about the electron microscope, we examined different samples amongst which the HepB sample prepared on August 16. The EM confirmed that indeed VLPs were formed.
29 August
- Miniprep of HepB PCR product in pSB1C3 backbone from colony PCR 20.Aug and HepB with his-tag (in pSB1C3 backbone) from colony PCR 20Aug to check again if there is an insert (since colony PCR is not working well sometimes)
-> the DNA concentration of the minipreps was very low!
- PCR check of the minipreps with both HepB primers as well as sequencing primers
-> some bands (with the right size) are visible for HepB with the his-tag and HepBwt (original HepB core protein) – but also in the same lane a second band (about the size of the backbone) - no conclusion can be made
30 August
- Inoculate sample 20 from 20July to make a new HepB+IPTG promoter biobrick – stock (cells did not grow)
-> did not grow
31 August
- again miniprep of HepB PCR product in pSB1C3 backbone from colony PCR 20.Aug and HepB with his-tag
-> this time the yield was higher
Hepatitis B outside modification
- 27 August (Kees)
• Whole plasmid PCR
Instead of using the general FW and Rev primers, the whole plasmid can be amplified alternatively. This allows a more controlled Tm and only 1 amplicon instead of 2 which have to be combined. The same steps 1-4 are still required.
First try with Phusion polymerase did not have any result.
- 29 August (Kees)
• Gradient PCR
A greadient pcr was performed using Taq polymerase. This can only determine wether the primers do work at a certain, yet for phusion undertermined temperature. Besides, Taq adds an A, causing a mutation in the final step. Therefore, the products can not be used, even if we accept the absence of proofreading in Taq.
• Phusion Polymerase PCR
After the proof was given that this first pcr step is possible, we use phusion to obtain usable product. The new annealing temperature is 63˚C, based upon the Fermentas calculation. The elongation time was set to 2min, which should allow elongation of the product at an elongation speed of 1k bp/min. Again, the pcr did not work. Therefore, new primers were ordered which can execute the whole pcr at once, including a TEV-protease sight.
Hepatitis B inside modification
27.August
- Gel extraction of PCR step 3 sample from 24.Aug
- 4th (final) step of the PCR reaction with the sample from the gel purification
-> this step seemed to work - the gel shows a band at the expected size slighly larger than the band from the 3rd step.
- Gel purification of the fragment of step 4 (H4)
28.August
- ligation of the final PCR product (HepB-insidecoil) in Bba_J04500 and transformation with Mach1
GFP modification
27.August
- 2nd try of the check and amplification of GFP-coil PCR product from 14.Aug
-> still no band visible for the GFP check/amplification
- Miniprep of GFP-coil in pSB1C3 backbone in JM109 (transformation from 20 August) to check again wheather there is an insert
29 August
- PCR check of the miniprep from 27. August for the GFP-coil insert (with GFP-coil primers as well as sequencing primers)
-> no bands visible on the gel - the plasmid does not contain the right insert
30. August
- 2nd try of the digestion, ligation and transformation of GFP-coil fragment into IPTG promoter backbone (Bba_J04500)
-> very little growth, also of the positive control - the transformation was not efficient! also there was a very low concentration of DNA after the purification (to purify the fragments from the restriction enzymes that could not be inactivated by heat) so the ligation might also have failed due to that
- Ligation of GFP-coil PCR product (not purified – sample from 13.Aug) with pJET
- Transformation of the ligation product with Mach1 (once with the direct ligation product and once with purified ligation product (PCR purification)
-> no colonies on the plate
31 August
- again miniprep of GFP-coil in pSB1C3 backbone in JM109 (transformation from 20 August) and HepB with his-tag
-> this time the yield was higher
CCMV Stability experiments
27th August (Mark)
- pH stability experiment with DLS
- pH range 4.5 - 7.5
- pH change was to sudden not measurable with DLS
PLRV
Because the expression of PLRV coat proteins was not successful in E. coli DH5α we transformed E. coli BL21 cells with the IPTG+RBS_PLRV_CP constructs (PLRV coat proteins downstream a IPTG inducible promoter plus ribosomal binding site). After expression, a SDS-PAGE gel was run but again no difference in protein expression was noticed.
 "
"