Team:Wageningen UR/Journal/week12
From 2012.igem.org
(→Lab work) |
(→Lab work) |
||
| (36 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
== Office work == | == Office work == | ||
| + | '''PLRV''' | ||
| + | |||
| + | This week we designed and ordered all the primers we needed. | ||
| + | [[File:primers.jpg]] | ||
== Lab work == | == Lab work == | ||
| - | + | '''General''' | |
| - | + | BBa_J04450 (RFP coding device) transformation, digestion check | |
| + | *BBa_J04450 was solubilized from the Standard registry parts plate I with 10µl MQ water | ||
| + | *Transformation of electrocompetent ''E.coli'' with the plasmid | ||
| + | *Plated on LB-agar with corresponding antibiotic (chloramphenicol) | ||
| + | *6 red-fluorescing colony were picked after overnight growth in 37°C | ||
| + | *Subsequent miniprep | ||
| + | |||
| + | {| class="wikitable" style="text-align: center" | ||
| + | |- style="font-style: italic" | ||
| + | |sample | ||
| + | |concentration (ng/µl) | ||
| + | |260/280 | ||
| + | |- | ||
| + | |A1 | ||
| + | |115.5 | ||
| + | |1.92 | ||
| + | |- | ||
| + | |C1 | ||
| + | |96.0 | ||
| + | |1.98 | ||
| + | |- | ||
| + | |C3 | ||
| + | |100.1 | ||
| + | |1.92 | ||
| + | |} | ||
| - | + | *Digestion check with EcoRI and SpeI | |
| - | + | [[File:RFP pSB1C3. digestion check.jpg|300px|center|thumb|''Figure 1: digestion BBa_J04450'']] | |
| - | + | ||
| - | + | ||
| - | + | ||
| + | Digestion of BBa_K1970- 22/38 | ||
| - | + | From the transformation plate of week 9 a new colony was grown and subsequently a miniprep was done. The yield was about 160 ng/µl. To check if the transformed E.coli contains the right plasmid a digestion check with the restriction enzymes BamHI and BgEII was done. | |
| + | [[File:Digest BBa K197022-38.jpg|500px|center|thumb|''Figure 2: digestion BBa_K197022/38'']] | ||
| - | ''' | + | ''' CCMV ''' |
| - | + | ''18th July'' (Hugo) | |
| - | + | <br/> | |
| - | + | First PCR on CCMV-Delta26 to create the CCMV_NEG construct. | |
| - | + | ||
| + | Phusion Mastermix for 4 + 1 = 5 samples: | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | ''' | + | {| class="wikitable" style="text-align: center" |
| + | |- style="font-style: italic" | ||
| + | |Substance | ||
| + | |Amount (µl) | ||
| + | |- | ||
| + | |H2O | ||
| + | |176.25 uL | ||
| + | |- | ||
| + | |5x Phusion Buffer | ||
| + | |50 uL | ||
| + | |- | ||
| + | |dNTP's | ||
| + | |5 uL | ||
| + | |- | ||
| + | |Phusion enzyme | ||
| + | |1.25 uL | ||
| + | |- | ||
| + | |Total | ||
| + | |232.5 uL | ||
| + | |} | ||
| + | |||
| + | Preparation samples: | ||
| + | |||
| + | |||
| + | {| class="wikitable" style="text-align: center" | ||
| + | |- style="font-style: italic" | ||
| + | |Substance | ||
| + | |Amount (µl) | ||
| + | |- | ||
| + | |Mastermix | ||
| + | |44 uL | ||
| + | |- | ||
| + | |DNA template | ||
| + | |1 uL | ||
| + | |- | ||
| + | |Forward Primer (10x diluted) | ||
| + | |2.5 uL | ||
| + | |- | ||
| + | |Reverse Primer (10x diluted) | ||
| + | |2.5 uL | ||
| + | |- | ||
| + | |Total | ||
| + | |50 uL | ||
| + | |} | ||
| + | |||
| + | PCR reaction: | ||
| + | |||
| + | {| class="wikitable" style="text-align: center" | ||
| + | |- style="font-style: italic" | ||
| + | |Step | ||
| + | |Time | ||
| + | |Temperature (°C) | ||
| + | |- | ||
| + | |Step 1 | ||
| + | |10 minutes | ||
| + | |98 °C | ||
| + | |- | ||
| + | |Step 2 | ||
| + | |30 seconds | ||
| + | |98 °C | ||
| + | |- | ||
| + | |Step 3 | ||
| + | |30 seconds | ||
| + | |58 °C | ||
| + | |- | ||
| + | |Step 4 | ||
| + | |30 seconds | ||
| + | |72 °C | ||
| + | |Return to step 2 (35 times) | ||
| + | |- | ||
| + | |Step 5 | ||
| + | |10 minutes | ||
| + | |72 °C | ||
| + | |- | ||
| + | |Step 6 | ||
| + | |As long as it takes | ||
| + | |4 °C | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | Gel: | ||
| + | |||
| + | {| class="wikitable" style="text-align: center" | ||
| + | |- style="font-style: italic" | ||
| + | |Lane | ||
| + | |Sample | ||
| + | |- | ||
| + | |Lane 1 | ||
| + | |Marker | ||
| + | |- | ||
| + | |Lane 2 | ||
| + | |Negative control: No template, same primers as duplo’s | ||
| + | |- | ||
| + | |Lane 3 | ||
| + | |Positive control: Delta26-Histag CCMV with Forward + Reverse (suffix) primer. | ||
| + | |- | ||
| + | |Lane 4 | ||
| + | |duplo 1: CCMV-Delta26 with Forward (Part 2) + Reverse (suffix) primer. | ||
| + | |- | ||
| + | |Lane 5 | ||
| + | |duplo 2: CCMV-Delta26 with Forward (Part 2) + Reverse (suffix) primer. | ||
| + | |} | ||
| + | |||
| + | [[File:Gel_18-07.tif|600px|center|thumb|''Figure 3: 1% agarose gel''</p>]] | ||
| + | |||
| + | ''19th July'' (Hugo) | ||
| + | <br/> | ||
| + | Second PCR on CCMV-Delta26. Before PCR was started, fragments of the day before were purified using a PCR purification kit. | ||
| + | <br/> | ||
| + | <br/> | ||
| + | ''20th July'' (Hugo) | ||
| + | <br/> | ||
| + | PCR repeat of 18-07. | ||
| + | <br/> | ||
| + | <br/> | ||
| + | |||
| + | |||
| + | ''' HepB ''' | ||
| + | |||
| + | * Digestion of the HepB core protein PCR product with pre- and suffix (from 11.July) and ligation into BBa_J04500 (an IPTG inducible promoter with RBS)as well as BBa_PSB1K3.ml (linearized plasmid backbone) | ||
| + | * Electro transformation of these constructs with DH5α | ||
| + | * Colony PCR (20 samples of the transformants containing HepB core protein + BBa_J04500; 10 samples of the transformants containing HepB core protein + BBa_PSB1K3.ml | ||
| + | -> The ligation and transformation of the HepB core protein into the linear backbone (BBa_PSB1K3.ml) was not successful | ||
| + | -> The colony PCR shows that we have 6 out of 20 colonies with the expected insert size (around 600bp) | ||
| + | |||
| + | |||
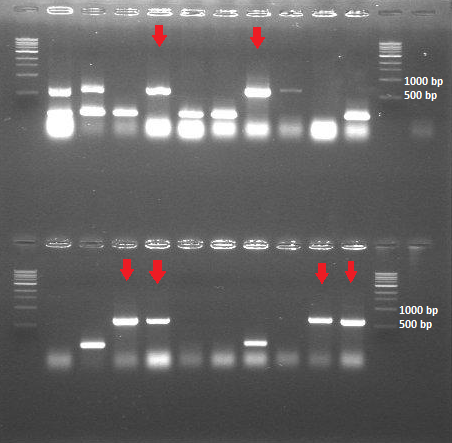
| + | [[File:ColonyPCR_20July.png|200px|center|thumb|<p align="justify">''Figure 4: Colony PCR 20 July - HepB in BBa_J04500'' </p>]] | ||
| + | |||
| - | |||
| - | |||
| - | |||
| - | |||
'''TuYV''' | '''TuYV''' | ||
| - | * Mach1 electrocompetent ''E. coli'' cells were transformed with last week's ligation mixtures. Results were unsatisfying. | + | * Mach1 electrocompetent ''E. coli'' cells were transformed with last week's ligation mixtures (TuYV coat protein and TuYV coat protein with his-tag). Results were unsatisfying. No colonies were showed on the sample plates. |
* 'Coat Protein + readthrough' gene amplicons were digested, ligated into pSB1K3 backbone and transformed into our electrocompetent Mach1 ''E. coli'' cells. After two attempts and a colony PCR, results were still unsatisfying. | * 'Coat Protein + readthrough' gene amplicons were digested, ligated into pSB1K3 backbone and transformed into our electrocompetent Mach1 ''E. coli'' cells. After two attempts and a colony PCR, results were still unsatisfying. | ||
| Line 59: | Line 205: | ||
After a two week quest for PLRV infected plant material, we obtained infected potato leafs from the Dutch General Inspection Service for Agricultural seed and seed potatoes. | After a two week quest for PLRV infected plant material, we obtained infected potato leafs from the Dutch General Inspection Service for Agricultural seed and seed potatoes. | ||
| - | + | ---- | |
| + | |||
| + | '''DLS boundary experiment''' | ||
| - | |||
| + | ''19th July'' (Mark) | ||
| + | <ul> | ||
| + | <li>DLS measurment: 20 runs, 45 seconds</li> | ||
| + | <ul> | ||
| + | <li>Changed 4 times to different vials to see if the vials are important to the measurement</li> | ||
| + | </ul> | ||
| + | </ul> | ||
---- | ---- | ||
[[https://2012.igem.org/Team:Wageningen_UR/Journal/week11 previous week]] [[https://2012.igem.org/Team:Wageningen_UR/Journal/week13 next week]] | [[https://2012.igem.org/Team:Wageningen_UR/Journal/week11 previous week]] [[https://2012.igem.org/Team:Wageningen_UR/Journal/week13 next week]] | ||
Latest revision as of 03:25, 27 September 2012
week 12: 16 july - 22 july
Office work
PLRV
This week we designed and ordered all the primers we needed.
Lab work
General
BBa_J04450 (RFP coding device) transformation, digestion check
- BBa_J04450 was solubilized from the Standard registry parts plate I with 10µl MQ water
- Transformation of electrocompetent E.coli with the plasmid
- Plated on LB-agar with corresponding antibiotic (chloramphenicol)
- 6 red-fluorescing colony were picked after overnight growth in 37°C
- Subsequent miniprep
| sample | concentration (ng/µl) | 260/280 |
| A1 | 115.5 | 1.92 |
| C1 | 96.0 | 1.98 |
| C3 | 100.1 | 1.92 |
- Digestion check with EcoRI and SpeI
Digestion of BBa_K1970- 22/38
From the transformation plate of week 9 a new colony was grown and subsequently a miniprep was done. The yield was about 160 ng/µl. To check if the transformed E.coli contains the right plasmid a digestion check with the restriction enzymes BamHI and BgEII was done.
CCMV
18th July (Hugo)
First PCR on CCMV-Delta26 to create the CCMV_NEG construct.
Phusion Mastermix for 4 + 1 = 5 samples:
| Substance | Amount (µl) |
| H2O | 176.25 uL |
| 5x Phusion Buffer | 50 uL |
| dNTP's | 5 uL |
| Phusion enzyme | 1.25 uL |
| Total | 232.5 uL |
Preparation samples:
| Substance | Amount (µl) |
| Mastermix | 44 uL |
| DNA template | 1 uL |
| Forward Primer (10x diluted) | 2.5 uL |
| Reverse Primer (10x diluted) | 2.5 uL |
| Total | 50 uL |
PCR reaction:
| Step | Time | Temperature (°C) | |
| Step 1 | 10 minutes | 98 °C | |
| Step 2 | 30 seconds | 98 °C | |
| Step 3 | 30 seconds | 58 °C | |
| Step 4 | 30 seconds | 72 °C | Return to step 2 (35 times) |
| Step 5 | 10 minutes | 72 °C | |
| Step 6 | As long as it takes | 4 °C |
Gel:
| Lane | Sample |
| Lane 1 | Marker |
| Lane 2 | Negative control: No template, same primers as duplo’s |
| Lane 3 | Positive control: Delta26-Histag CCMV with Forward + Reverse (suffix) primer. |
| Lane 4 | duplo 1: CCMV-Delta26 with Forward (Part 2) + Reverse (suffix) primer. |
| Lane 5 | duplo 2: CCMV-Delta26 with Forward (Part 2) + Reverse (suffix) primer. |
19th July (Hugo)
Second PCR on CCMV-Delta26. Before PCR was started, fragments of the day before were purified using a PCR purification kit.
20th July (Hugo)
PCR repeat of 18-07.
HepB
- Digestion of the HepB core protein PCR product with pre- and suffix (from 11.July) and ligation into BBa_J04500 (an IPTG inducible promoter with RBS)as well as BBa_PSB1K3.ml (linearized plasmid backbone)
- Electro transformation of these constructs with DH5α
- Colony PCR (20 samples of the transformants containing HepB core protein + BBa_J04500; 10 samples of the transformants containing HepB core protein + BBa_PSB1K3.ml
-> The ligation and transformation of the HepB core protein into the linear backbone (BBa_PSB1K3.ml) was not successful -> The colony PCR shows that we have 6 out of 20 colonies with the expected insert size (around 600bp)
TuYV
- Mach1 electrocompetent E. coli cells were transformed with last week's ligation mixtures (TuYV coat protein and TuYV coat protein with his-tag). Results were unsatisfying. No colonies were showed on the sample plates.
- 'Coat Protein + readthrough' gene amplicons were digested, ligated into pSB1K3 backbone and transformed into our electrocompetent Mach1 E. coli cells. After two attempts and a colony PCR, results were still unsatisfying.
PLRV
After a two week quest for PLRV infected plant material, we obtained infected potato leafs from the Dutch General Inspection Service for Agricultural seed and seed potatoes.
DLS boundary experiment
19th July (Mark)
- DLS measurment: 20 runs, 45 seconds
- Changed 4 times to different vials to see if the vials are important to the measurement
 "
"