Team:University College London/Module 2/Modelling
From 2012.igem.org
(→Reactions taking place in the model) |
Erinoerton (Talk | contribs) (→Reactions taking place in the model) |
||
| (26 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
{{:Team:University_College_London/templates/module2menu}} | {{:Team:University_College_London/templates/module2menu}} | ||
| - | == | + | == Curli synthesis == |
| - | Our cell model for aggregation describes the pathways through which | + | |
| + | Our cell model for aggregation describes the pathways through which curli cluster coding genes interact to make up up curli fibrils which are produced on detection of microplastics. This particular curli cluster (BBa_K540000) contains five genes that later on interact and make up curli fibrils, however, we are looking into synthesis and interaction of only two of them: CsgA and CsgB due to complex nature of the system. | ||
| + | |||
| + | == Aim == | ||
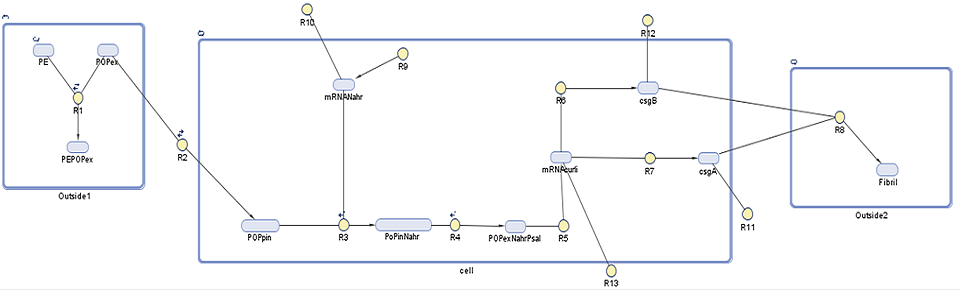
| + | Using this we aim to find out to which extent is curli fibrils' production is dependent on the persistent organic pollutant presence in the surrounding environment. In addition to that we would like to estimate how much fibrils are produced over time.The following diagram shows the genes, molecules and reaction involved in the synthesis of the curli fibrils, on which further description is provided below. | ||
[[File:aggnet.png]] | [[File:aggnet.png]] | ||
| - | As with our [[Team:University_College_London/Module_3/Modelling|degradation model]], this SimBiology model is divided into three compartments: | + | == Description of the model == |
| + | |||
| + | As with our [[Team:University_College_London/Module_3/Modelling|degradation model]], this SimBiology model is divided into three compartments, with each DNA species and reaction explained in further detail below: | ||
| - | 1. '''Outside1:''' in this compartment we see the association, adherence, and dissociation of persistent organic pollutants (POPs) from polyethylene (PE) (R1). We rely on the POPs to induce our | + | 1. '''Outside1:''' in this compartment we see the association, adherence, and dissociation of persistent organic pollutants (POPs) from polyethylene (PE) (R1). We rely on the POPs to induce our curli synthesis system. As it will be described later on, POPs have a tendency to adhere to the plastic 10<sup>5</sup> to 10<sup>6</sup> times more than reamin in the ocean. As curli synthesis is induced by the POPs this means that curli synthesis will be the specificity to our system. |
| - | 2. '''Cell:''' NahR is a constitutively produced mRNA product (R9) of which around 3% degrades (R10). When POP diffuses into the cell (R2), it forms a complex with intact NahR (R3) which then binds to the pSal promoter (R4) to induce the curli gene cluster (R5). This starts translation of five proteins which make up curli fibrils. Here we focus on those controlled by the CsgAB operon, CsgA (R7) and CsgB (R6), as these are the most important in curli synthesis as described on our | + | 2. '''Cell:''' NahR is a constitutively produced mRNA product (R9) of which around 3% degrades (R10). When POP diffuses into the cell (R2), it forms a complex with intact NahR (R3) which then binds to the pSal promoter (R4) to induce the curli gene cluster (R5). This starts translation of five proteins which make up curli fibrils. Here we focus on those controlled by the CsgAB operon, CsgA (R7) and CsgB (R6), as these are the most important in curli synthesis as described on our [https://2012.igem.org/Team:University_College_London/Module_2 research page]. The CsgA and CsgB that do not degrade (R11, R12) diffuse out of the cell. |
| Line 31: | Line 37: | ||
| POPex || 0.0 || Persistent organic pollutants (ex = extracellular) that are not adhered to plastic surface | | POPex || 0.0 || Persistent organic pollutants (ex = extracellular) that are not adhered to plastic surface | ||
|- | |- | ||
| - | | PEPOPex || 9.24E-5 || Persistent organic pollutants (ex = extracellular) that are adhered to the plastic surface<sup> | + | | PEPOPex || 9.24E-5 || Persistent organic pollutants (ex = extracellular) that are adhered to the plastic surface<sup>6</sup> |
|- | |- | ||
| POPin || 0.5 || Persistent organic pollutants (in = intracellular) assumed from <i>E. coli</i> membrane permeability <sup>4</sup> | | POPin || 0.5 || Persistent organic pollutants (in = intracellular) assumed from <i>E. coli</i> membrane permeability <sup>4</sup> | ||
| Line 41: | Line 47: | ||
| POPinNahRpSal || 0.0 || Complex of the above molecule and pSal (promoter that induces laccase transcription) | | POPinNahRpSal || 0.0 || Complex of the above molecule and pSal (promoter that induces laccase transcription) | ||
|- | |- | ||
| - | | mRNACurli || 0.0 || Polycistronic mRNA as it codes for more than one protein, in reality curli cluster contains five or more proteins | + | | mRNACurli || 0.0 || Polycistronic mRNA as it codes for more than one protein, in reality curli cluster contains five or more proteins but as mentioned previously we are concentrating only on the synthesis of CsgA and CsgA therefore we assume that only two genes are present in this polycistronic mRNA. |
|- | |- | ||
| CsgA || 0.0 || One of the polypeptides that is coded for in curli cluster, it is a structural component secreted in subunits outside of the cell | | CsgA || 0.0 || One of the polypeptides that is coded for in curli cluster, it is a structural component secreted in subunits outside of the cell | ||
| Line 56: | Line 62: | ||
! Number !! Reaction !! Reaction rate (molecules/sec) !! Notes & Assumptions | ! Number !! Reaction !! Reaction rate (molecules/sec) !! Notes & Assumptions | ||
|- | |- | ||
| - | | R1 || PE + POPex ↔ PEPOPex || Forward: 1000 <br /> Backward: 1 || Pops have | + | | R1 || PE + POPex ↔ PEPOPex || Forward: 1000 <br /> Backward: 1 || Pops have 100000 to 1000000 times greater tendency to adhere to plastic than float free in the ocean<sup>6</sup> |
|- | |- | ||
| R2 || POPex ↔ POPin || Forward: 0.6 <br /> Backward: 0.4 || Rate based on membrane permeability<sup>4</sup> and diffusion gradient | | R2 || POPex ↔ POPin || Forward: 0.6 <br /> Backward: 0.4 || Rate based on membrane permeability<sup>4</sup> and diffusion gradient | ||
|- | |- | ||
| - | | R3 || POPin + mRNA.Nahr | + | | R3 || POPin + mRNA.Nahr ↔ POPin.mRNA.Nahr || Forward: 1 <br /> Backward: 0.0001 || Based on the assumption that the chemical structure/size of POPs is similar to salycilate<sup>7</sup>. Salycilate binds to the NahR mRNA product, which complex then binds to the pSal promoter. |
|- | |- | ||
| - | | R4 || POPinmRNANahr | + | | R4 || POPinmRNANahr ↔ POPinmRNANahr.Psal || Forward: 78200 <br /> Backward: 0.191 <sup>10</sup> || NahR to pSal binding based on the assumption that POP-NahR binding has no effect on NahR-pSal binding |
|- | |- | ||
| R5 || POPexmRNANahr.Psal → mRNAcurli || 0.054 || Transcription rate of curli cluster in molecules/sec (for cluster size 1500bp<sup>11</sup>, transcription rate in E.coli 80bp/sec<sup>9</sup>) | | R5 || POPexmRNANahr.Psal → mRNAcurli || 0.054 || Transcription rate of curli cluster in molecules/sec (for cluster size 1500bp<sup>11</sup>, transcription rate in E.coli 80bp/sec<sup>9</sup>) | ||
| Line 70: | Line 76: | ||
| R7 || mRNAcurli → CsgA || 0.13 || Translation rate of CsgA in molecules/sec (for CsgA size 151aa<sup>13</sup>, translation rate in E.coli 20aa/sec<sup>9</sup>) | | R7 || mRNAcurli → CsgA || 0.13 || Translation rate of CsgA in molecules/sec (for CsgA size 151aa<sup>13</sup>, translation rate in E.coli 20aa/sec<sup>9</sup>) | ||
|- | |- | ||
| - | | R8 || CsgB + CsgA -> fibril || | + | | R8 || CsgB + CsgA -> fibril || 0.003 || See calculations below |
|- | |- | ||
| - | | R9 || 0 | + | | R9 || 0 ↔ mRNA.Nahr || Forward: 0.088 <br /> Backward: 0.6 || Transcription rate of NahR in molecules/sec (for NahR size 909 bp<sup>8</sup>, transcription rate in E.coli 80bp/sec<sup>9</sup>) under constitutive promoter control |
|- | |- | ||
| R10 || mRNA.Nahr → 0 || 0.03 || Degradation rate of NahR mRNA product<sup>14</sup> | | R10 || mRNA.Nahr → 0 || 0.03 || Degradation rate of NahR mRNA product<sup>14</sup> | ||
| Line 80: | Line 86: | ||
| R12 || CsgB → 0 || 0.03 || Degradation rate of CsgB <sup>14</sup> must be taken into account due to suboptimal conditions | | R12 || CsgB → 0 || 0.03 || Degradation rate of CsgB <sup>14</sup> must be taken into account due to suboptimal conditions | ||
|} | |} | ||
| + | |||
| + | |||
| + | We calculated the reaction rate of R8 (CsgB + CsgA -> fibril) as follows: | ||
| + | |||
| + | The average length of an amino acid is 0.8nm<sup>15</sup>. The length of CsgA is 82 amino acids<sup>16</sup>, so a non-folded CsgA protein is 65.6 nm in length. CsgA's tertiary structure is folded into 5<sup>17</sup> so the length of a CsgA protein in a curli is 13.1 nm. A curli is several micrometers long (~3000nm)<sup>18</sup> so there are around 299 CsgA molecules per curli. The production of CsgA is the limiting step in the production of curli fibrils (compared to production of CsgB) so the forward rate for this reaction is 1/299. | ||
== Results == | == Results == | ||
| - | + | The following graph shows that fibrils are produced at a rate of around 1 every three hours. | |
| - | + | [[File:agggraph3.jpg]] | |
== References == | == References == | ||
1. Goldstein M, Rosenberg M, Cheng L (2012) Increased oceanic microplastic debris enhances oviposition in an endemic pelagic insect. <i>Biology Letters</i> 10.1098 | 1. Goldstein M, Rosenberg M, Cheng L (2012) Increased oceanic microplastic debris enhances oviposition in an endemic pelagic insect. <i>Biology Letters</i> 10.1098 | ||
| - | 2. Andrady AL (2011) Microplastics in the marine environment. Marine Pollution Bulletin 62: 1596-1605 | + | 2. Andrady AL (2011) Microplastics in the marine environment. <i>Marine Pollution Bulletin</i> 62: 1596-1605 |
| - | + | 4. Kay J, Koivusalo M, Ma X, Wohland T, Grinstein S (2012) Phosphatidylserine Dynamics in Cellular Membranes. <i>Molecular Biology of the Cell</i> | |
| - | + | ||
| - | 4. | + | |
5. Nenninger AA, Robinson LS, Hammer ND, Epstein EA, Badtke MP, Hultgren SJ, Chapman MR (2011) CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. <i>Mol Microbiol</i> 81: 486-499 | 5. Nenninger AA, Robinson LS, Hammer ND, Epstein EA, Badtke MP, Hultgren SJ, Chapman MR (2011) CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. <i>Mol Microbiol</i> 81: 486-499 | ||
| - | 6. Mato Y, Isobe T, Takada H, Kanehiro H, Ohtake C, Kaminuma T (2001) Plastic Resin Pellets as a Transport Medium for Toxic Chemicals in the Marine Environment. Environ. Sci. Technol. 35: 318-324 | + | 6. Mato Y, Isobe T, Takada H, Kanehiro H, Ohtake C, Kaminuma T (2001) Plastic Resin Pellets as a Transport Medium for Toxic Chemicals in the Marine Environment. <i>Environ. Sci. Technol.</i> 35: 318-324 |
7. https://2011.igem.org/Team:Peking_S/project/wire/harvest | 7. https://2011.igem.org/Team:Peking_S/project/wire/harvest | ||
| Line 116: | Line 125: | ||
14. Kushner S (2002) mRNA Decay in <i>Escherichia coli</i> Comes of Age. <i>J Bacteriol.</i> 184: 4658-4665 | 14. Kushner S (2002) mRNA Decay in <i>Escherichia coli</i> Comes of Age. <i>J Bacteriol.</i> 184: 4658-4665 | ||
| - | + | 15. http://en.wikibooks.org/wiki/Cell_Biology/Introduction/Cell_size | |
| + | |||
| + | 16. http://bacteria.ensembl.org/b_subtilis/Transcript/Summary?g=EBBACG00000004082;r=Chromosome:228066-228314;t=EBBACT00000004090 | ||
| + | |||
| + | 17. Shu Q, Crick S, Pinkner J, Ford B, Hultgren S, Frieden C (2012) The E. coli CsgB nucleator of curli assembles to β-sheet oligomers that alter the CsgA fibrillization mechanism. Proceedings of the National Academy of Sciences of the United States of America 109: 6502-6507. DOI: 10.1073/pnas.1204161109 | ||
| + | |||
| + | 18. Pitkanen M, Honkalampi U, von Wright A, Sneck A, Hentze H-P, Sievanen J, Hiltunen J, Hellen EKO (2010) Nanofibrillar cellulose - assessment of cytotoxic and genotoxic properties <i>in vitro</i>. International conference on nanotechnology for the forest products industry | ||
Latest revision as of 14:57, 26 September 2012
Contents |
Module 2: Aggregation
Description | Design | Construction | Characterisation | Shear Device | Modelling | Results | Conclusions
Curli synthesis
Our cell model for aggregation describes the pathways through which curli cluster coding genes interact to make up up curli fibrils which are produced on detection of microplastics. This particular curli cluster (BBa_K540000) contains five genes that later on interact and make up curli fibrils, however, we are looking into synthesis and interaction of only two of them: CsgA and CsgB due to complex nature of the system.
Aim
Using this we aim to find out to which extent is curli fibrils' production is dependent on the persistent organic pollutant presence in the surrounding environment. In addition to that we would like to estimate how much fibrils are produced over time.The following diagram shows the genes, molecules and reaction involved in the synthesis of the curli fibrils, on which further description is provided below.
Description of the model
As with our degradation model, this SimBiology model is divided into three compartments, with each DNA species and reaction explained in further detail below:
1. Outside1: in this compartment we see the association, adherence, and dissociation of persistent organic pollutants (POPs) from polyethylene (PE) (R1). We rely on the POPs to induce our curli synthesis system. As it will be described later on, POPs have a tendency to adhere to the plastic 105 to 106 times more than reamin in the ocean. As curli synthesis is induced by the POPs this means that curli synthesis will be the specificity to our system.
2. Cell: NahR is a constitutively produced mRNA product (R9) of which around 3% degrades (R10). When POP diffuses into the cell (R2), it forms a complex with intact NahR (R3) which then binds to the pSal promoter (R4) to induce the curli gene cluster (R5). This starts translation of five proteins which make up curli fibrils. Here we focus on those controlled by the CsgAB operon, CsgA (R7) and CsgB (R6), as these are the most important in curli synthesis as described on our research page. The CsgA and CsgB that do not degrade (R11, R12) diffuse out of the cell.
3. Outside2: Outside of the cell the polymerization of CsgA by CsgB takes place (R8) to make a curli fibril.
Species
| Species | Initial value (molecules) | Notes & Assumptions |
|---|---|---|
| PE | 0.044 | Polyethylene found in North Pacific Gyre (value per cubic metre)1,2 |
| POPex | 0.0 | Persistent organic pollutants (ex = extracellular) that are not adhered to plastic surface |
| PEPOPex | 9.24E-5 | Persistent organic pollutants (ex = extracellular) that are adhered to the plastic surface6 |
| POPin | 0.5 | Persistent organic pollutants (in = intracellular) assumed from E. coli membrane permeability 4 |
| mRNANahR | 0.0 | NahR mRNA product |
| POPinNahR | 0.0 | Complex of the above two molecules |
| POPinNahRpSal | 0.0 | Complex of the above molecule and pSal (promoter that induces laccase transcription) |
| mRNACurli | 0.0 | Polycistronic mRNA as it codes for more than one protein, in reality curli cluster contains five or more proteins but as mentioned previously we are concentrating only on the synthesis of CsgA and CsgA therefore we assume that only two genes are present in this polycistronic mRNA. |
| CsgA | 0.0 | One of the polypeptides that is coded for in curli cluster, it is a structural component secreted in subunits outside of the cell |
| CsgB | 0.0 | One of the polypeptides that is coded for in curli cluster, it is secreted outside of the cell allowing polymerization of CsgA |
| Fibril | 0.0 | Result of CsgA and CsgB interaction |
Reactions taking place in the model
| Number | Reaction | Reaction rate (molecules/sec) | Notes & Assumptions |
|---|---|---|---|
| R1 | PE + POPex ↔ PEPOPex | Forward: 1000 Backward: 1 | Pops have 100000 to 1000000 times greater tendency to adhere to plastic than float free in the ocean6 |
| R2 | POPex ↔ POPin | Forward: 0.6 Backward: 0.4 | Rate based on membrane permeability4 and diffusion gradient |
| R3 | POPin + mRNA.Nahr ↔ POPin.mRNA.Nahr | Forward: 1 Backward: 0.0001 | Based on the assumption that the chemical structure/size of POPs is similar to salycilate7. Salycilate binds to the NahR mRNA product, which complex then binds to the pSal promoter. |
| R4 | POPinmRNANahr ↔ POPinmRNANahr.Psal | Forward: 78200 Backward: 0.191 10 | NahR to pSal binding based on the assumption that POP-NahR binding has no effect on NahR-pSal binding |
| R5 | POPexmRNANahr.Psal → mRNAcurli | 0.054 | Transcription rate of curli cluster in molecules/sec (for cluster size 1500bp11, transcription rate in E.coli 80bp/sec9) |
| R6 | mRNAcurli → CsgB | 0.13 | Translation rate of CsgB in molecules/sec (for CsgB size 151aa12, translation rate in E.coli 20aa/sec9) |
| R7 | mRNAcurli → CsgA | 0.13 | Translation rate of CsgA in molecules/sec (for CsgA size 151aa13, translation rate in E.coli 20aa/sec9) |
| R8 | CsgB + CsgA -> fibril | 0.003 | See calculations below |
| R9 | 0 ↔ mRNA.Nahr | Forward: 0.088 Backward: 0.6 | Transcription rate of NahR in molecules/sec (for NahR size 909 bp8, transcription rate in E.coli 80bp/sec9) under constitutive promoter control |
| R10 | mRNA.Nahr → 0 | 0.03 | Degradation rate of NahR mRNA product14 |
| R11 | CsgA → 0 | 0.03 | Degradation rate of CsgA 14 must be taken into account due to suboptimal conditions |
| R12 | CsgB → 0 | 0.03 | Degradation rate of CsgB 14 must be taken into account due to suboptimal conditions |
We calculated the reaction rate of R8 (CsgB + CsgA -> fibril) as follows:
The average length of an amino acid is 0.8nm15. The length of CsgA is 82 amino acids16, so a non-folded CsgA protein is 65.6 nm in length. CsgA's tertiary structure is folded into 517 so the length of a CsgA protein in a curli is 13.1 nm. A curli is several micrometers long (~3000nm)18 so there are around 299 CsgA molecules per curli. The production of CsgA is the limiting step in the production of curli fibrils (compared to production of CsgB) so the forward rate for this reaction is 1/299.
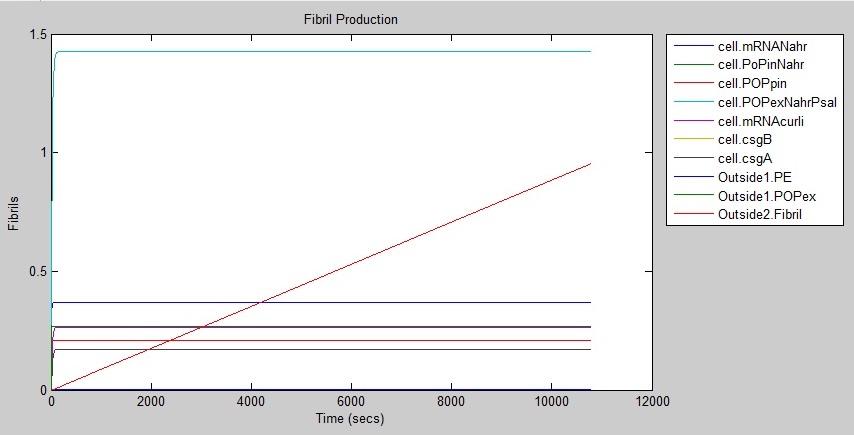
Results
The following graph shows that fibrils are produced at a rate of around 1 every three hours.
References
1. Goldstein M, Rosenberg M, Cheng L (2012) Increased oceanic microplastic debris enhances oviposition in an endemic pelagic insect. Biology Letters 10.1098
2. Andrady AL (2011) Microplastics in the marine environment. Marine Pollution Bulletin 62: 1596-1605
4. Kay J, Koivusalo M, Ma X, Wohland T, Grinstein S (2012) Phosphatidylserine Dynamics in Cellular Membranes. Molecular Biology of the Cell
5. Nenninger AA, Robinson LS, Hammer ND, Epstein EA, Badtke MP, Hultgren SJ, Chapman MR (2011) CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. Mol Microbiol 81: 486-499
6. Mato Y, Isobe T, Takada H, Kanehiro H, Ohtake C, Kaminuma T (2001) Plastic Resin Pellets as a Transport Medium for Toxic Chemicals in the Marine Environment. Environ. Sci. Technol. 35: 318-324
7. https://2011.igem.org/Team:Peking_S/project/wire/harvest
8. http://www.xbase.ac.uk/genome/azoarcus-sp-bh72/NC_008702/azo2419;nahR1/viewer
9. http://kirschner.med.harvard.edu/files/bionumbers/fundamentalBioNumbersHandout.pdf
10. Park H, Lim W, Shin H (2005) In vitro binding of purified NahR regulatory protein with promoter Psal. Biochimica et Biophysica Acta 1775: 247-255
11. Shala AA, Restrepo S, Gonzalez Barrios AF (2011) A network model for biofilm development in Escherichia coli K-12. Theoretical Biology and Medical Modelling 8: 34 doi:10.1186/1742-4682-8-34
12. http://www.ecogene.org//?q=gene/EG12621
13. http://www.ecogene.org/?q=gene/EG11489
14. Kushner S (2002) mRNA Decay in Escherichia coli Comes of Age. J Bacteriol. 184: 4658-4665
15. http://en.wikibooks.org/wiki/Cell_Biology/Introduction/Cell_size
17. Shu Q, Crick S, Pinkner J, Ford B, Hultgren S, Frieden C (2012) The E. coli CsgB nucleator of curli assembles to β-sheet oligomers that alter the CsgA fibrillization mechanism. Proceedings of the National Academy of Sciences of the United States of America 109: 6502-6507. DOI: 10.1073/pnas.1204161109
18. Pitkanen M, Honkalampi U, von Wright A, Sneck A, Hentze H-P, Sievanen J, Hiltunen J, Hellen EKO (2010) Nanofibrillar cellulose - assessment of cytotoxic and genotoxic properties in vitro. International conference on nanotechnology for the forest products industry
 "
"