Team:University College London/BioBricks
From 2012.igem.org
Rwilkinson (Talk | contribs) (→Module 5: Salt Tolerance) |
(→Module 4: Buoyancy) |
||
| (5 intermediate revisions not shown) | |||
| Line 43: | Line 43: | ||
== Module 4: Buoyancy == | == Module 4: Buoyancy == | ||
| - | + | [[File:BOUYANCYGFP.png|center|900px]] | |
| - | '''Figure 4:''' The image above depicts the circuit for Module 4 (Buoyancy). The Starvation Promoter (BBa_K118011) drives expression of | + | |
| + | |||
| + | [[File:BOUY.png|center|900px]] | ||
| + | |||
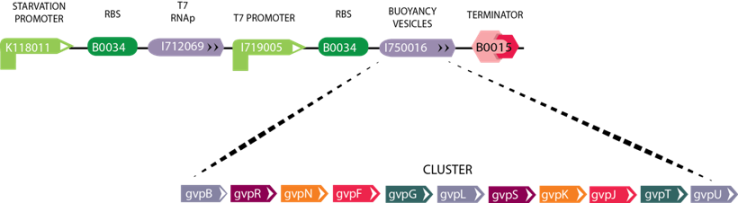
| + | '''Figure 4:''' The image above depicts the circuit for Module 4 (Buoyancy). The Starvation Promoter (BBa_K118011) drives expression of theT7 RNA polymerase when the environmental glucose concentration is low, then T7 RNA polymerase produced binds the T7 promoter resulting in the expression of GFP. As a future vision we will include the Gas Vesicle Cluster instead of the reporter gene. The Gas Vesicle Cluster selected originates from ''Bacillus Megaterium'', and induces buoyancy in the cell. | ||
For more information on how this circuit functions, please visit our [[Team:University_College_London/Module_4|Buoyancy Description Subpage]] | For more information on how this circuit functions, please visit our [[Team:University_College_London/Module_4|Buoyancy Description Subpage]] | ||
| + | |||
| + | Gas Vesicle Cluster | ||
== Module 5: Salt Tolerance == | == Module 5: Salt Tolerance == | ||
| Line 60: | Line 66: | ||
== Module: Containment == | == Module: Containment == | ||
| - | <html><div align="center"><img src="https://static.igem.org/mediawiki/2012/ | + | <html><div align="center"><img src="https://static.igem.org/mediawiki/2012/f/f9/UclContainment.png" alt="Containment" /></div></html> |
Latest revision as of 03:07, 27 September 2012
Contents |
BioBricks
BioBricks submitted to the Registry
<groupparts>iGEM012 University_College_London</groupparts>
Module 1: Plastic Detection

Figure 1: The figure above shows our genetic circuit for the Detection Module. The Constitutive Promoter (BBa_J23119) drives constant expression of the NahR protein BBa_K228004 (modified - see below). NahR is a transcriptional activator, which is activated in response to salicylate molecules. Our proxy for plastic - Organic Pollutants - have a salicylate-like domain, and are therefore capable of activating NahR. Activated NahR binds the pSal promoter (Figure 2) thus activating the circuit for Module 2 (Aggregation).
Modified BBa_K228004: The BBa_K228004 BioBrick must be modified as it combines both the NahR protein and the pSal promoter. We wish to seperate these.
Please see our Detection Description Subpage for more details on this Module.
Module 2: Aggregation

Figure 2: The Figure above illustrates our circuit for Module 2 (Aggregation). The pSal promoter BBa_K228004 (modified - see below) which triggers the Aggregation circuit is turned on when bound by activated NahR protein from Module 1. Once the pSal promoter is activated, it drives expression of the curli cluster of genes (BBa_K729003). The individuals genes of the cluster are shown radiating from BBa_K540000.
Modified BBa_K228004: The BBa_K228004 BioBrick must be modified as it combines both the NahR protein and the pSal promoter. We wish to seperate these.
BBa_K729003: This BioBrick is modified from BBa_K540000, which has a cobalt promoter.
For more information on how this circuit is triggered, and the individual roles of the Curli genes, please visit our Aggregation Description Subpage
Module 3: Plastic Degradation

Figure 3: The image above shows the circuit for Module 3 (Degradation). The Constitutive Promoter (BBa_J23119) drives constant expression of the Laccase BioBrick (BBa_K729002). The Laccase gene we intend to use originates from a particular bacterial strain, and is capable of Polyethylene Degradation.
For more information on how this circuit functions, please visit our Degradation Description Subpage
Module 4: Buoyancy
Figure 4: The image above depicts the circuit for Module 4 (Buoyancy). The Starvation Promoter (BBa_K118011) drives expression of theT7 RNA polymerase when the environmental glucose concentration is low, then T7 RNA polymerase produced binds the T7 promoter resulting in the expression of GFP. As a future vision we will include the Gas Vesicle Cluster instead of the reporter gene. The Gas Vesicle Cluster selected originates from Bacillus Megaterium, and induces buoyancy in the cell.
For more information on how this circuit functions, please visit our Buoyancy Description Subpage
Gas Vesicle Cluster
Module 5: Salt Tolerance

Figure 5: The image above illustrates the circuit design for Module 5 (Salt Tolerance). The Constitutive Promoter (BBa_J23119) will drive constant expression of our novel BioBrick IrrE, which confers salt resistance onto E. Coli.
For more information on how this circuit functions, please visit our Salt Tolerance Description Subpage
Module: Containment

Figure 6: The above image shows the genetic circuits for our threefold Containment module. Plasmid A carries the three toxin genes - Holin/Endolysin (BBa_K729009), Colicin E3 (BBa_K729005), and EcoRI (BBa_K729006). Plasmid B carries the antitoxins Anti-Holin (BBa_K729010), Colicin E3 Immuntiy (BBa_K729008), and Methytransferase Ecori. Plasmid A and B work in conjunction with one another to prevent Horizontal Gene Transfer by conjugation. Plasmid C carries a periplasmic nuclease (BBa_K279004) driven by the constitutive promoter BBa_J23119.
For more information on how this circuit functions, please visit our Containment Description Subpage
 "
"