Team:UNAM Genomics Mexico/Results/AND
From 2012.igem.org
| (46 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Template:Team:UNAM_Genomics_Mexico/webhtml| content= | {{:Template:Team:UNAM_Genomics_Mexico/webhtml| content= | ||
| - | + | __NOTOC__ | |
<br /> | <br /> | ||
| - | <center><h1>'''AND'''</h1></center> | + | <center><h1>'''AND's Results'''</h1></center> |
<br /> | <br /> | ||
| + | <center> | ||
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | ||
| + | <tr> | ||
| + | <td id="rightcolumn2" align= "center"><br />[[File:UnamgenomicsANDcadmio.png | 200px |link=Team:UNAM_Genomics_Mexico/Results/AND#CzrA-ArsR_AND_GATE]]<br /><br /><p>'''AND Heavy Metals'''</p></td> | ||
| + | <td id="leftcolumn3" align= "center"><br />[[File:UnamgenomicsANDarabinosa.png | 200px |link=Team:UNAM_Genomics_Mexico/Results/AND#ARABINOSE-XYLOSE_AND_GATE]]<p>'''AND Sugar''''</p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | <br/> | ||
| + | <br/> | ||
== '''CzrA-ArsR AND GATE''' == | == '''CzrA-ArsR AND GATE''' == | ||
| Line 10: | Line 20: | ||
'''HEAVY-METAL AND'''<br/> | '''HEAVY-METAL AND'''<br/> | ||
<br/> | <br/> | ||
| - | This construct was designed to function as a logic gate, an AND to be specific. This is due to the way the CzrA-ArsR promoter was designed. We used a system that sensed heavy metals in Bacillus subtilis, which was originally designed by the iGEM Newcastle team 2009 ( | + | This construct was designed to function as a logic gate, an AND to be specific. This is due to the way the CzrA-ArsR promoter was designed. We used a system that sensed heavy metals in Bacillus subtilis, which was originally designed by the iGEM Newcastle team 2009 ([https://2009.igem.org/Team:Newcastle/Project#Cadmium_Sensing Newcastle University iGEM team 2009]). This system consists of a fused promoter, which includes both ArsR and CzrA binding sites. Arar and CzrA are metal sensing repressors. They both respond to cadmium, however silver, arsenic, or copper induces ArsR and zinc, cobalt, or nickel induces CzrA as well (Moore CM, Helmann JD. Metal ion homeostasis in Bacillus subtilis. Curr Opin Microbiol. 2005 Apr;8(2):188-95.). If we use two different metals, specific for each repressor we will have an AND gate. Besides Newcastle’s 2009 design, we designed two different fused promoters with the same binding sites but in different order to try different combinations that could make the system more efficient. <br/> |
<br/> | <br/> | ||
<br/> | <br/> | ||
| - | To obtain our final construct we required the following Biological Parts:<br/> | + | '''To obtain our final construct we required the following Biological Parts''':<br/> |
<br/> | <br/> | ||
<br/> | <br/> | ||
'''Obtained from the registry:'''<br/> | '''Obtained from the registry:'''<br/> | ||
| - | BBa_K143001 (AmyE 5’)<br/> | + | [http://partsregistry.org/Part:BBa_K143001 BBa_K143001 (AmyE 5’)]<br/> |
| - | BBa_K143002 (AmyE 3’)<br/> | + | [http://partsregistry.org/Part:BBa_K143002 BBa_K143002] (AmyE 3’)<br/> |
| - | BBa_E1010 (RFP)<br/> | + | [http://partsregistry.org/Part:BBa_E1010 BBa_E1010 (RFP)]<br/> |
| - | BBa_B0014 (Terminator)<br/> | + | [http://partsregistry.org/Part:BBa_B0014 BBa_B0014 (Terminator)]<br/> |
| - | BBa_C0179 (LasR)<br/> | + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_C0179 BBa_C0179 (LasR)]<br/> |
| - | Synthesis Products:<br/> | + | <br/> |
| + | <br/> | ||
| + | '''Synthesis Products:'''<br/> | ||
CzrA-AsR 99<br/> | CzrA-AsR 99<br/> | ||
CzrA-AsR 98<br/> | CzrA-AsR 98<br/> | ||
| Line 28: | Line 40: | ||
RBS-CI<br/> | RBS-CI<br/> | ||
<br/> | <br/> | ||
| - | |||
| - | |||
<br/> | <br/> | ||
| + | '''Obtained from Margarita Salas Ph.D.’s Group:'''<br/> | ||
| + | P4 from phage phi29 from plasmid pRMn25.<br/> | ||
<br/> | <br/> | ||
| - | |||
<br/> | <br/> | ||
| - | + | '''Omega cassette from plasmid pHP45Ω.'''<br/> | |
<br/> | <br/> | ||
| + | We designed the following primers to add the RBS site to BBa_C0179, BBa_E100 and P4.<br/> | ||
<br/> | <br/> | ||
| - | LASR_2.0_seq_registry<br/> | + | '''LASR_2.0_seq_registry'''<br/> |
UPPER 5'-3'<br/> | UPPER 5'-3'<br/> | ||
PREFIJO+RBS+ESPACIADOR+LASR<br/> | PREFIJO+RBS+ESPACIADOR+LASR<br/> | ||
| - | GTTTCTTCGAATTCGCGGCCGCTTCTAGAG AAAGGTGGTGAA TACTAG atggccttggttgac<br/> | + | GTTTCTTCGAATTCGCGGCCGCTTCTAGAG AAAGGTGGTGAA TACTAG atggccttggttgac<br/><br/> |
LOWER 5'-3'<br/> | LOWER 5'-3'<br/> | ||
SUFIJO+LASR_<br/> | SUFIJO+LASR_<br/> | ||
GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTA ttattagagagtaat<br/> | GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTA ttattagagagtaat<br/> | ||
<br/> | <br/> | ||
| - | RFP<br/> | + | '''RFP'''<br/> |
UPPER 5'-3'<br/> | UPPER 5'-3'<br/> | ||
PREFIX+RBS+SPACER+RFP <br/> | PREFIX+RBS+SPACER+RFP <br/> | ||
| - | |||
GTTTCTTCGAATTCGCGGCCGCTTCTAGAG AAAGGTGGTGAA TACTAG ATGGCTTCCTCCGAA<br/> | GTTTCTTCGAATTCGCGGCCGCTTCTAGAG AAAGGTGGTGAA TACTAG ATGGCTTCCTCCGAA<br/> | ||
<br/> | <br/> | ||
LOWER 5'-3'<br/> | LOWER 5'-3'<br/> | ||
SUFIX+RFP<br/> | SUFIX+RFP<br/> | ||
| - | |||
GTTTCTTCCTGCAGCGGCCGCTACTAGTA TTATTAAGCACCGGT<br/> | GTTTCTTCCTGCAGCGGCCGCTACTAGTA TTATTAAGCACCGGT<br/> | ||
<br/> | <br/> | ||
| - | + | '''P4''' <br/> | |
| - | + | ||
| - | P4 <br/> | + | |
PREFIX+RBS+SPACER+P4 <br/> | PREFIX+RBS+SPACER+P4 <br/> | ||
UPPER<br/> | UPPER<br/> | ||
| - | |||
GTTTCTTCGAATTCGCGGCCGCTTCTAGAG AAAGGTGGTGAA TACTAG ATGCCTAAAACACAA<br/> | GTTTCTTCGAATTCGCGGCCGCTTCTAGAG AAAGGTGGTGAA TACTAG ATGCCTAAAACACAA<br/> | ||
<br/> | <br/> | ||
SUFIX+P4<br/> | SUFIX+P4<br/> | ||
LOWER 5'-3'<br/> | LOWER 5'-3'<br/> | ||
| - | |||
GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTA CTACACCATACTTTT<br/> | GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTA CTACACCATACTTTT<br/> | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
This metal AND team had to build the following construct:<br/> | This metal AND team had to build the following construct:<br/> | ||
| - | + | <center> | |
| - | < | + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> |
| + | <tr> | ||
| + | <td id="rightcolumn2" align= "center"><br />[[File:UnamgenomicsAndmetals1.png| x90px]]</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | |||
The other AND (pBad/pXyl) team was in charge of building this part. <br/> | The other AND (pBad/pXyl) team was in charge of building this part. <br/> | ||
<br/> | <br/> | ||
| - | So we | + | |
| + | <center> | ||
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | ||
| + | <tr> | ||
| + | <td id="leftcolumn3" align= "center"><br />[[File:UnamgenomicsresultsOr3.png| x80px]]</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | |||
| + | |||
| + | '''So we obtain the final product:'''<br/> | ||
<br/> | <br/> | ||
| + | After the Regional Jamboree, we continued working very hard, and our work has paid off, these our new results: | ||
| + | Amy,98,P4,CI,RFP,TT<br/> | ||
| + | Amy99,P4,CI,RFP,TT<br/> | ||
| + | Also we feel very proud and excited because we finally have one complete AND gate =D !!!!<br/> | ||
| + | AMY,97,P4,CI,RFP,TT,OMEGA,AMY3<br/> | ||
<br/> | <br/> | ||
| - | + | <center> | |
| - | + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | |
| - | + | <tr> | |
| - | + | <td id="rightcolumn2" align= "center"><br />[[File:UnamgenomicsConstruccion1.png| x120px]]</td> | |
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
<br/> | <br/> | ||
| - | + | ||
| + | <center> | ||
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | ||
| + | <tr> | ||
| + | <td id="rightcolumn2" align= "center"><br />[[File:UnamgenomicsmexicoandResultspartsmetal.png| 800px]]<p>'''AmyE 5’_ArsR/CzrA 97-99, P4_CI, RFP_Terminator, P4_CI_RFP_Terminator and AmyE 5’_CzrA/ArsR 99-97_P4CI_RFP_Terminator'''</p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | |||
| + | ::::::::::::::::::::::::::::::::::::::[[File:UnamgenomcisUp.png|right | 120px |link=Team:UNAM_Genomics_Mexico/Results/AND]] | ||
<br/> | <br/> | ||
| - | |||
<br/> | <br/> | ||
<br/> | <br/> | ||
| - | + | ||
| - | + | == '''ARABINOSE-XYLOSE AND GATE''' == | |
| - | + | ||
| - | + | ||
| - | + | ||
<br/> | <br/> | ||
| - | + | '''SWEET AND''' | |
<br/> | <br/> | ||
| - | |||
| - | |||
<br/> | <br/> | ||
This construct was designed to function as a AND logic gate. This is due to the way the pBad/pXyl promoter was designed. We used a system that sensed l-arabinose in E.coli which was originally designed by Amelia Hardjasa and used by iGEM09_British_Columbia1,2. In the absence of arabinose, the repressor protein AraC (BBa_C0080) binds to the AraI1 operator site of pBAD and the upstream operator site AraO2, blocking transcription1. In the presence of arabinose, AraC binds to it and changes its conformation such that it interacts with the AraI1 and AraI2 operator sites, permitting transcription1. We also used a promoter inducible by xylose that has been designed for high expression in B.subtilis which was originally designed by James Chappell and used by iGEM08_Imperial_College. Xylose does not induce the promoter xylose directly, but requires the transcriptional regulator XylR (BBa_K143036). Our system consists in a fused promoter which includes both AraC and XylR binding sites. AraC and XylR are l-arabinose and xylose sensing, respectively, repressors. In this way, if we use these two inputs, each specific for each repressor we will have an AND gate.<br/> | This construct was designed to function as a AND logic gate. This is due to the way the pBad/pXyl promoter was designed. We used a system that sensed l-arabinose in E.coli which was originally designed by Amelia Hardjasa and used by iGEM09_British_Columbia1,2. In the absence of arabinose, the repressor protein AraC (BBa_C0080) binds to the AraI1 operator site of pBAD and the upstream operator site AraO2, blocking transcription1. In the presence of arabinose, AraC binds to it and changes its conformation such that it interacts with the AraI1 and AraI2 operator sites, permitting transcription1. We also used a promoter inducible by xylose that has been designed for high expression in B.subtilis which was originally designed by James Chappell and used by iGEM08_Imperial_College. Xylose does not induce the promoter xylose directly, but requires the transcriptional regulator XylR (BBa_K143036). Our system consists in a fused promoter which includes both AraC and XylR binding sites. AraC and XylR are l-arabinose and xylose sensing, respectively, repressors. In this way, if we use these two inputs, each specific for each repressor we will have an AND gate.<br/> | ||
| Line 103: | Line 136: | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| - | Obtained from the Registry:<br/> | + | '''Obtained from the Registry:'''<br/> |
| - | BBa_K143001 (AmyE 5’)<br/> | + | [http://partsregistry.org/Part:BBa_K143001 BBa_K143001 (AmyE 5’)]<br/> |
| - | BBa_K143002 (AmyE 3’)<br/> | + | [http://partsregistry.org/Part:BBa_K143002 BBa_K143002 (AmyE 3’)]<br/> |
| - | BBa_E1010 (RFP)<br/> | + | [http://partsregistry.org/Part:BBa_E1010 BBa_E1010 (RFP)]<br/> |
| - | BBa_B0014 ( | + | [http://partsregistry.org/Part:BBa_B0014 BBa_B0014 (Terminator)]<br/> |
| - | BBa_C0080 (AraC)<br/> | + | [http://partsregistry.org/wiki/index.php/Part:BBa_C0080 BBa_C0080 (AraC)]<br/> |
| - | Synthesis Products:<br/> | + | <br/> |
| + | '''Synthesis Products:'''<br/> | ||
pBad/pXyl promoter<br/> | pBad/pXyl promoter<br/> | ||
RBS XylR<br/> | RBS XylR<br/> | ||
| - | Obtained from Margarita Salas Ph.D.’s Group:<br/> | + | |
| + | '''Obtained from Margarita Salas Ph.D.’s Group:'''<br/> | ||
A3 from phage phi29 from plasmid pFRC54.<br/> | A3 from phage phi29 from plasmid pFRC54.<br/> | ||
<br/> | <br/> | ||
| - | Omega cassette from plasmid pHP45Ω.<br/> | + | '''Omega cassette from plasmid pHP45Ω.'''<br/> |
<br/> | <br/> | ||
We designed the following primers to add the RBS site to BBa_E1010 (RFP) and BBa_C0080 (AraC):<br/> | We designed the following primers to add the RBS site to BBa_E1010 (RFP) and BBa_C0080 (AraC):<br/> | ||
| - | RFP<br/> | + | '''RFP'''<br/> |
UPPER 5'-3'<br/> | UPPER 5'-3'<br/> | ||
PREFIX+RBS+SPACER+RFP <br/> | PREFIX+RBS+SPACER+RFP <br/> | ||
| Line 126: | Line 161: | ||
GTTTCTTCCTGCAGCGGCCGCTACTAGTA TTATTAAGCACCGGT<br/> | GTTTCTTCCTGCAGCGGCCGCTACTAGTA TTATTAAGCACCGGT<br/> | ||
<br/> | <br/> | ||
| - | ARAC without LVA<br/> | + | '''ARAC without LVA'''<br/> |
UPPER 5'-3'<br/> | UPPER 5'-3'<br/> | ||
PREFIX+RBS+SPACER+ARAC<br/> | PREFIX+RBS+SPACER+ARAC<br/> | ||
| Line 136: | Line 171: | ||
We also designed the following primers to obtain A3 from phage phi29 from plasmid pFRC54 and Omega cassette from plasmid pHP45Ω:<br/> | We also designed the following primers to obtain A3 from phage phi29 from plasmid pFRC54 and Omega cassette from plasmid pHP45Ω:<br/> | ||
<br/> | <br/> | ||
| - | A3 <br/> | + | '''A3''' <br/> |
UPPER 5'-3'<br/> | UPPER 5'-3'<br/> | ||
PREFIX+A3<br/> | PREFIX+A3<br/> | ||
| Line 145: | Line 180: | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| - | OMEGA CASSETTE <br/> | + | '''OMEGA CASSETTE''' <br/> |
PREFIX+OMEGA CASSETTE(45 bp)<br/> | PREFIX+OMEGA CASSETTE(45 bp)<br/> | ||
5' GTTTCTTCGAATTCGCGGCCGCTTCTAGAG CCGGGGATCCGGTGA 3'<br/> | 5' GTTTCTTCGAATTCGCGGCCGCTTCTAGAG CCGGGGATCCGGTGA 3'<br/> | ||
| Line 154: | Line 189: | ||
<br/> | <br/> | ||
This arabinose-xylose AND team had to build the following constructs:<br/> | This arabinose-xylose AND team had to build the following constructs:<br/> | ||
| + | |||
| + | <center> | ||
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | ||
| + | <tr> | ||
| + | <td id="leftcolumn3" align= "center"><br />[[File:UnamgenomicsresutsAmypbadpxy.png | x90px]]<br /><br /><p></p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td id="leftcolumn3" align= "center"><br />[[File:UnamgenomicsresultsP4lasr.png | x90px]]<p></p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td id="leftcolumn3" align= "center"><br />[[File:UnamgenomicsResultsabisugar1.png | x90px]]<p></p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| + | The other AND (heavy metals) team was in charge of building this part.<br/> | ||
<br/> | <br/> | ||
| + | |||
| + | <center> | ||
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | ||
| + | <tr> | ||
| + | <td id="rightcolumn2" align= "center"><br />[[File:UnamgenomicsRfgstop.png| x90px]]<p>'''RFP_Terminator'''</p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
<br/> | <br/> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br/> | <br/> | ||
So we could obtain the final product:<br/> | So we could obtain the final product:<br/> | ||
<br/> | <br/> | ||
| - | <br/> | + | <center> |
| - | < | + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> |
| - | < | + | <tr> |
| + | <td id="leftcolumn3" align= "center"><br />[[File:Unamgenomicsdeepconstruccion 2.png| x115px]]<p></p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
<br/> | <br/> | ||
What we have up now is:<br/> | What we have up now is:<br/> | ||
<br/> | <br/> | ||
| - | AmyE | + | |
| + | <center> | ||
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | ||
| + | <tr> | ||
| + | <td id="leftcolumn3" align= "center"><br />[[File:UnamgenomicsResultssugar.png| 850px]]<p>'''AmyE 5’_pBad/pXyl, pVeg_RBS_XylR, Omega cassette_AmyE 3’, RBS_AraC_Omega cassette_AmyE 3’'''</p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| - | + | We are working hard to obtain these last two constructs in order to finish it:<br/> | |
<br/> | <br/> | ||
| + | |||
| + | <center> | ||
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | ||
| + | <tr> | ||
| + | <td id="leftcolumn3" align= "center"><br />[[File:UnamgenomicsresultsSugarfalta.png| 850px]]<p>'''pVeg_RBS_XylR_RBS_AraC_Omega cassette_AmyE 3’, AmyE 5’_pBad/pXyl_RBS_P4/LasR_RBS_CI_RBS_RFP_Double Terminador'''</p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| - | + | ::::::::::::::::::::::::::::::::::::::[[File:UnamgenomcisUp.png|right | 120px |link=Team:UNAM_Genomics_Mexico/Results/AND]] | |
| - | < | + | |
| - | < | + | '''Please see our wetlab notebook in the clicking the following image:''' |
| - | <br/> | + | |
| - | + | <center> | |
| - | <br/> | + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> |
| - | < | + | <tr> |
| - | < | + | <td id="rightcolumn2" align= "center"><br />[[File:UnamgenomicsANDcadmio.png | 200px |link=Team:UNAM_Genomics_Mexico/Notebook/ANDMetal]]<br /><br /><p>'''AND Heavy Metals Notebook'''</p></td> |
| - | + | ||
| - | < | + | <td id="leftcolumn3" align= "center"><br />[[File:UnamgenomicsANDarabinosa.png | 200px |link=Team:UNAM_Genomics_Mexico/Notebook/ANDSugar]]<p>'''AND Sugar Notebook''''</p></td> |
| - | + | </tr> | |
| - | < | + | </table> |
| - | < | + | </center> |
| - | + | == '''References''' == | |
<br/> | <br/> | ||
| - | |||
[1] Schlief, R. (2000). Regulation of the L-arabinose operon of Escherichia coli. Trends in Genetics. 16(12):559-565. <br/> | [1] Schlief, R. (2000). Regulation of the L-arabinose operon of Escherichia coli. Trends in Genetics. 16(12):559-565. <br/> | ||
[2] Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, and Keasling JD. (2001). Homogeneous expression of the PBAD promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology. 147(12):3241-7. <br/> | [2] Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, and Keasling JD. (2001). Homogeneous expression of the PBAD promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology. 147(12):3241-7. <br/> | ||
Latest revision as of 00:36, 27 October 2012

AND's Results
 AND Heavy Metals |
 AND Sugar' |
CzrA-ArsR AND GATE
HEAVY-METAL AND
This construct was designed to function as a logic gate, an AND to be specific. This is due to the way the CzrA-ArsR promoter was designed. We used a system that sensed heavy metals in Bacillus subtilis, which was originally designed by the iGEM Newcastle team 2009 (Newcastle University iGEM team 2009). This system consists of a fused promoter, which includes both ArsR and CzrA binding sites. Arar and CzrA are metal sensing repressors. They both respond to cadmium, however silver, arsenic, or copper induces ArsR and zinc, cobalt, or nickel induces CzrA as well (Moore CM, Helmann JD. Metal ion homeostasis in Bacillus subtilis. Curr Opin Microbiol. 2005 Apr;8(2):188-95.). If we use two different metals, specific for each repressor we will have an AND gate. Besides Newcastle’s 2009 design, we designed two different fused promoters with the same binding sites but in different order to try different combinations that could make the system more efficient.
To obtain our final construct we required the following Biological Parts:
Obtained from the registry:
BBa_K143001 (AmyE 5’)
BBa_K143002 (AmyE 3’)
BBa_E1010 (RFP)
BBa_B0014 (Terminator)
BBa_C0179 (LasR)
Synthesis Products:
CzrA-AsR 99
CzrA-AsR 98
CzrA-AsR 97
RBS-CI
Obtained from Margarita Salas Ph.D.’s Group:
P4 from phage phi29 from plasmid pRMn25.
Omega cassette from plasmid pHP45Ω.
We designed the following primers to add the RBS site to BBa_C0179, BBa_E100 and P4.
LASR_2.0_seq_registry
UPPER 5'-3'
PREFIJO+RBS+ESPACIADOR+LASR
GTTTCTTCGAATTCGCGGCCGCTTCTAGAG AAAGGTGGTGAA TACTAG atggccttggttgac
LOWER 5'-3'
SUFIJO+LASR_
GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTA ttattagagagtaat
RFP
UPPER 5'-3'
PREFIX+RBS+SPACER+RFP
GTTTCTTCGAATTCGCGGCCGCTTCTAGAG AAAGGTGGTGAA TACTAG ATGGCTTCCTCCGAA
LOWER 5'-3'
SUFIX+RFP
GTTTCTTCCTGCAGCGGCCGCTACTAGTA TTATTAAGCACCGGT
P4
PREFIX+RBS+SPACER+P4
UPPER
GTTTCTTCGAATTCGCGGCCGCTTCTAGAG AAAGGTGGTGAA TACTAG ATGCCTAAAACACAA
SUFIX+P4
LOWER 5'-3'
GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTA CTACACCATACTTTT
This metal AND team had to build the following construct:
 |
The other AND (pBad/pXyl) team was in charge of building this part.
 |
So we obtain the final product:
After the Regional Jamboree, we continued working very hard, and our work has paid off, these our new results:
Amy,98,P4,CI,RFP,TT
Amy99,P4,CI,RFP,TT
Also we feel very proud and excited because we finally have one complete AND gate =D !!!!
AMY,97,P4,CI,RFP,TT,OMEGA,AMY3
 |
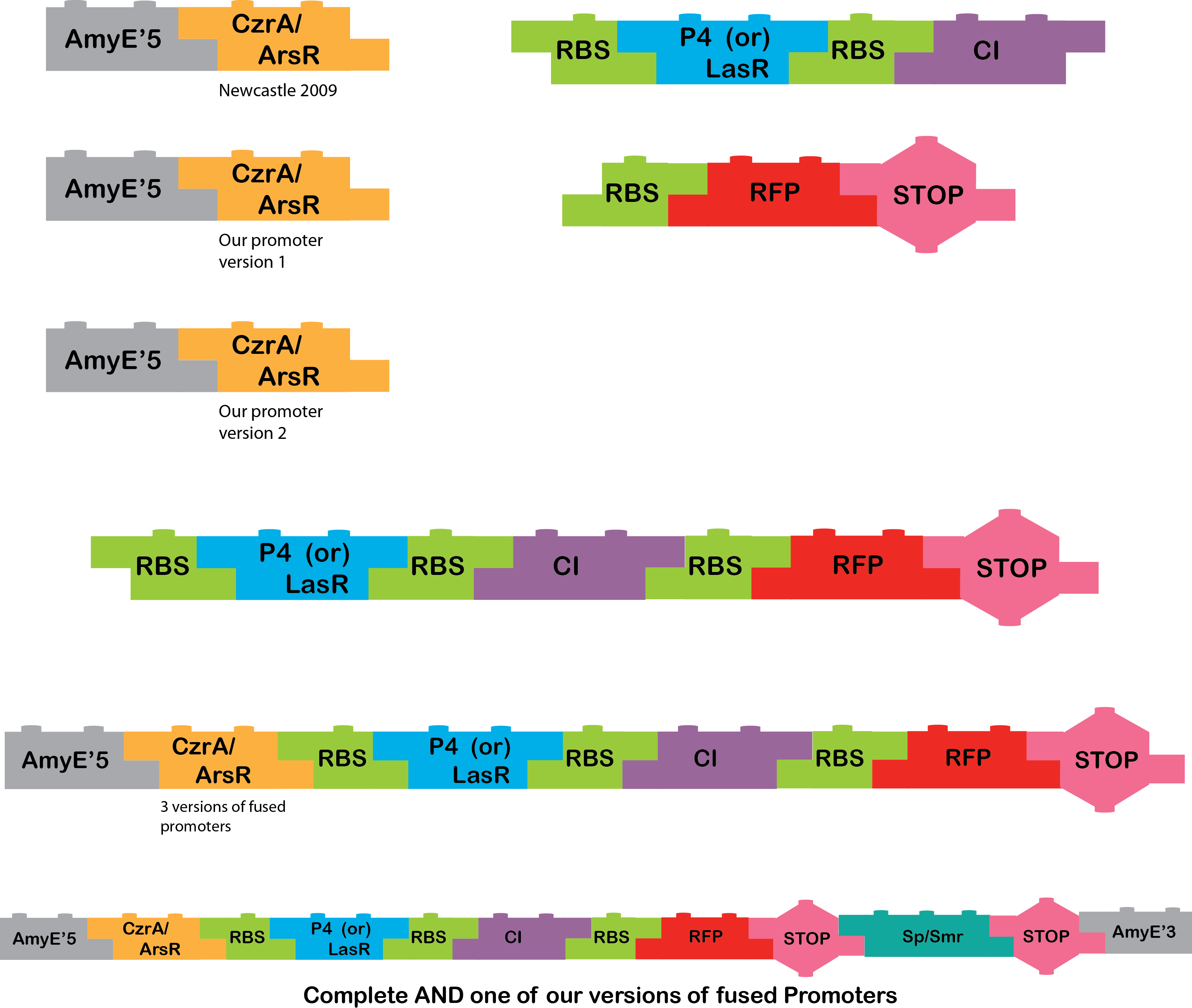 AmyE 5’_ArsR/CzrA 97-99, P4_CI, RFP_Terminator, P4_CI_RFP_Terminator and AmyE 5’_CzrA/ArsR 99-97_P4CI_RFP_Terminator |
ARABINOSE-XYLOSE AND GATE
SWEET AND
This construct was designed to function as a AND logic gate. This is due to the way the pBad/pXyl promoter was designed. We used a system that sensed l-arabinose in E.coli which was originally designed by Amelia Hardjasa and used by iGEM09_British_Columbia1,2. In the absence of arabinose, the repressor protein AraC (BBa_C0080) binds to the AraI1 operator site of pBAD and the upstream operator site AraO2, blocking transcription1. In the presence of arabinose, AraC binds to it and changes its conformation such that it interacts with the AraI1 and AraI2 operator sites, permitting transcription1. We also used a promoter inducible by xylose that has been designed for high expression in B.subtilis which was originally designed by James Chappell and used by iGEM08_Imperial_College. Xylose does not induce the promoter xylose directly, but requires the transcriptional regulator XylR (BBa_K143036). Our system consists in a fused promoter which includes both AraC and XylR binding sites. AraC and XylR are l-arabinose and xylose sensing, respectively, repressors. In this way, if we use these two inputs, each specific for each repressor we will have an AND gate.
To obtain our final construct we required the following Biological Parts:
Obtained from the Registry:
BBa_K143001 (AmyE 5’)
BBa_K143002 (AmyE 3’)
BBa_E1010 (RFP)
BBa_B0014 (Terminator)
BBa_C0080 (AraC)
Synthesis Products:
pBad/pXyl promoter
RBS XylR
Obtained from Margarita Salas Ph.D.’s Group:
A3 from phage phi29 from plasmid pFRC54.
Omega cassette from plasmid pHP45Ω.
We designed the following primers to add the RBS site to BBa_E1010 (RFP) and BBa_C0080 (AraC):
RFP
UPPER 5'-3'
PREFIX+RBS+SPACER+RFP
GTTTCTTCGAATTCGCGGCCGCTTCTAGAG AAAGGTGGTGAA TACTAG ATGGCTTCCTCCGAA
LOWER 5'-3'
SUFIX+RFP
GTTTCTTCCTGCAGCGGCCGCTACTAGTA TTATTAAGCACCGGT
ARAC without LVA
UPPER 5'-3'
PREFIX+RBS+SPACER+ARAC
GTTTCTTCGAATTCGCGGCCGCTTCTAGAG AAAGGTGGTGAA TACTAG ATGGCTGAAGCGCAA
LOWER 5'-3'
SUFIX+ARAC
GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTA CAACTTGACGGCTAC
We also designed the following primers to obtain A3 from phage phi29 from plasmid pFRC54 and Omega cassette from plasmid pHP45Ω:
A3
UPPER 5'-3'
PREFIX+A3
5' GTTTCTTCGAATTCGCGGCCGCTTCTAGAG taactttttgcaaga 3'
LOWER 5'-3'
SUFIX+A3
5'GTTTCTTCCTGCAGCGGCCGCTACTAGTA ctacttaattatacc 3'
OMEGA CASSETTE
PREFIX+OMEGA CASSETTE(45 bp)
5' GTTTCTTCGAATTCGCGGCCGCTTCTAGAG CCGGGGATCCGGTGA 3'
SUFIX+OMEGA CASSETTE (44 bp)
5' GTTTCTTCCTGCAGCGGCCGCTACTAGTA CCGGGGATCCGGTGA 3'
This arabinose-xylose AND team had to build the following constructs:
 |
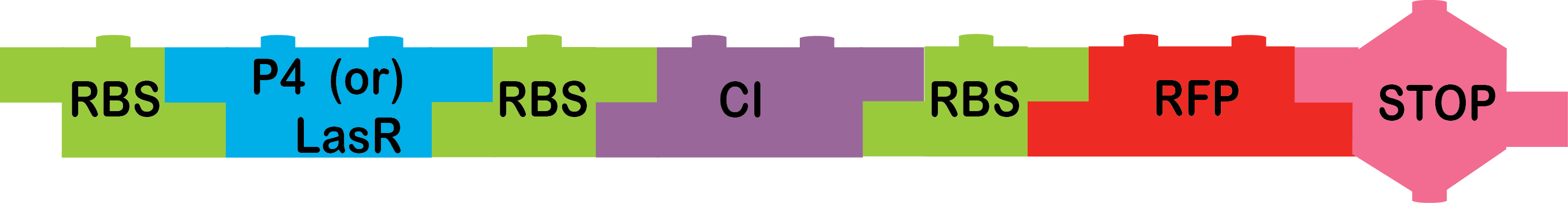 |
 |
The other AND (heavy metals) team was in charge of building this part.
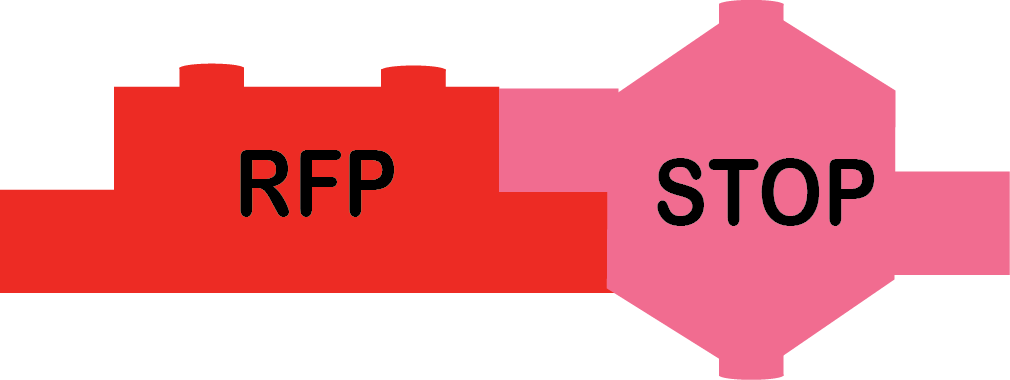 RFP_Terminator |
So we could obtain the final product:
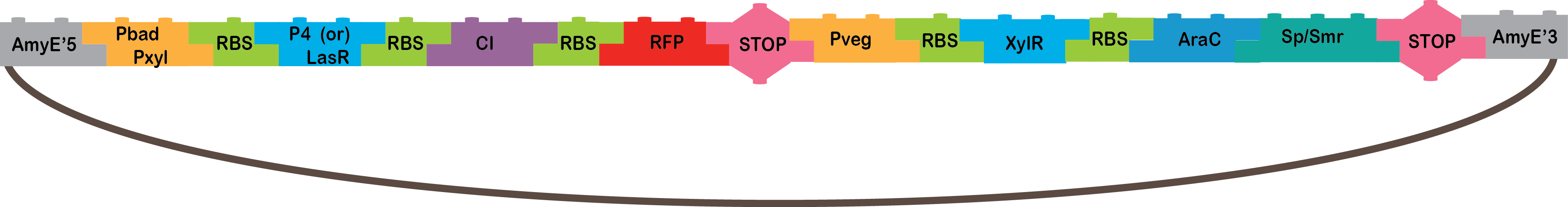 |
What we have up now is:
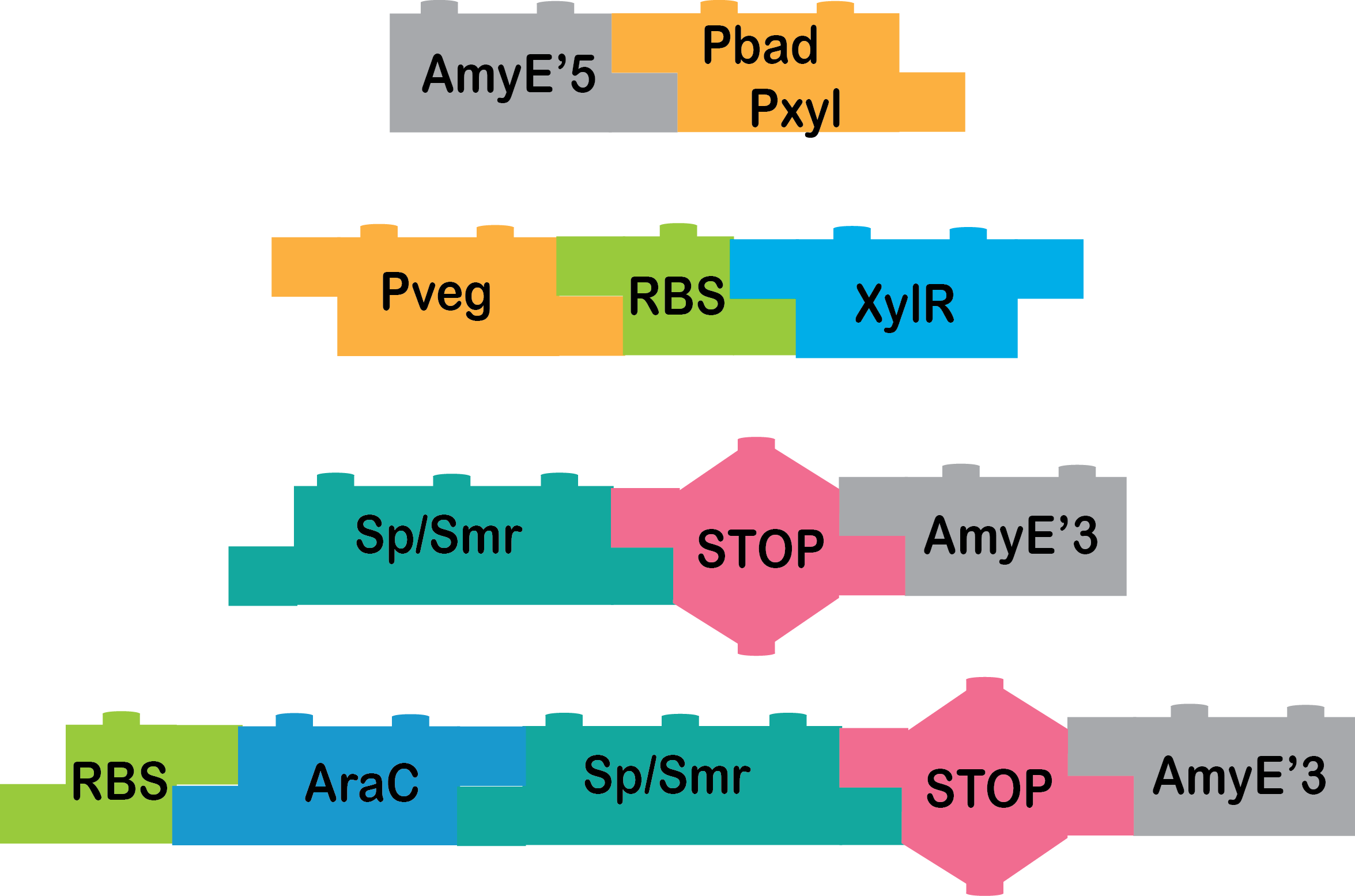 AmyE 5’_pBad/pXyl, pVeg_RBS_XylR, Omega cassette_AmyE 3’, RBS_AraC_Omega cassette_AmyE 3’ |
We are working hard to obtain these last two constructs in order to finish it:
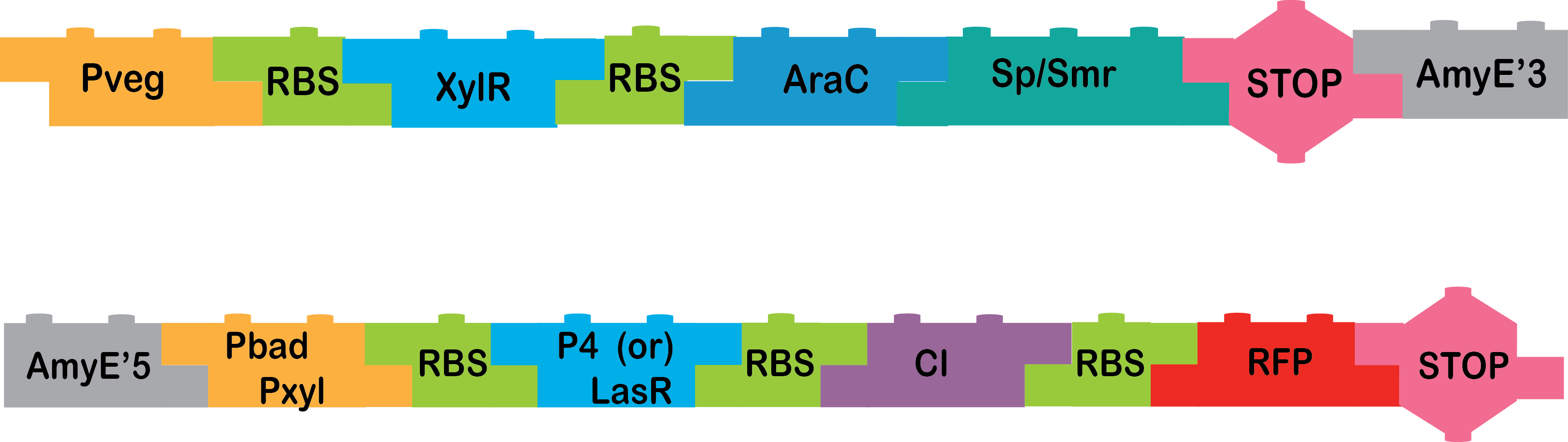 pVeg_RBS_XylR_RBS_AraC_Omega cassette_AmyE 3’, AmyE 5’_pBad/pXyl_RBS_P4/LasR_RBS_CI_RBS_RFP_Double Terminador |
Please see our wetlab notebook in the clicking the following image:
 AND Heavy Metals Notebook |
 AND Sugar Notebook' |
References
[1] Schlief, R. (2000). Regulation of the L-arabinose operon of Escherichia coli. Trends in Genetics. 16(12):559-565.
[2] Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, and Keasling JD. (2001). Homogeneous expression of the PBAD promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology. 147(12):3241-7.
 "
"







