Team:UC Chile2/Cyanolux/Constructs
From 2012.igem.org
(Difference between revisions)
| (4 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{ | + | {{UC_Chile4}} |
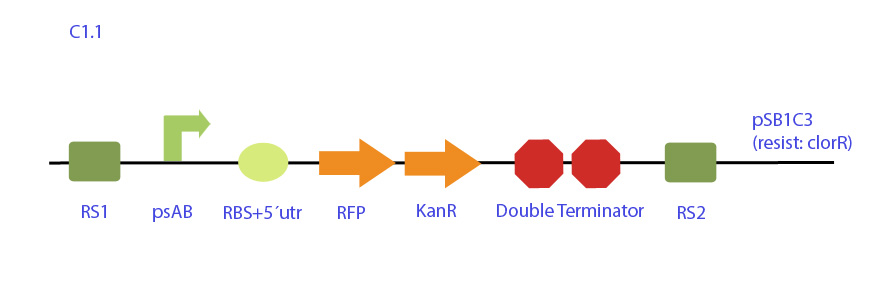
| - | [[File: | + | [[File:UC_Chile-C1.1.jpg|center]] |
| - | + | <p> | |
| - | <p | + | Integrative plasmid designed for the expression of RFP under synechocystis psbAB promoter also with a kanamycin resistance cassette for selection in cyanobacteria. The construct is flanked by neutral recombination sites RS1 and RS2, homologous to synechocystis chromosome.</p> |
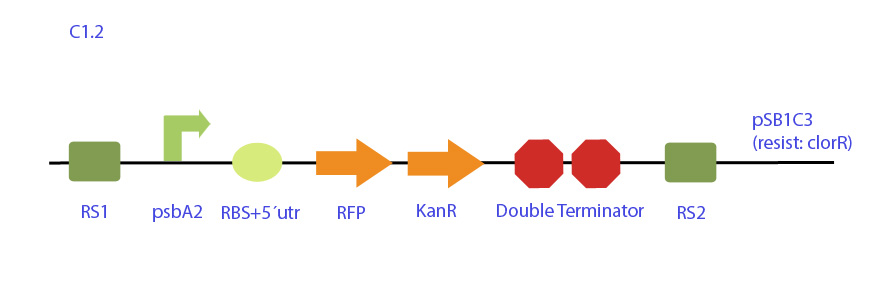
| - | Integrative plasmid designed for the expression of RFP under synechocystis psbAB promoter also with a kanamycin resistance cassette for selection in cyanobacteria. The construct is flanked by neutral recombination sites RS1 and RS2, homologous to synechocystis chromosome. | + | [[File:UC_Chile-C1.2.jpg|center]] |
| - | + | <p>C1.2 | |
| - | [[File: | + | |
| - | + | ||
| - | + | ||
| - | <p | + | |
This integrative plasmid is designed for the expression of RFP under synechocystis psaA2 promoter with a kanamycin resistance cassette for selection in cyanobacteria. The construct is flanked by neutral recombination sites RS1 and RS2, homologous to synechocystis chromosome. | This integrative plasmid is designed for the expression of RFP under synechocystis psaA2 promoter with a kanamycin resistance cassette for selection in cyanobacteria. The construct is flanked by neutral recombination sites RS1 and RS2, homologous to synechocystis chromosome. | ||
| - | + | </p> | |
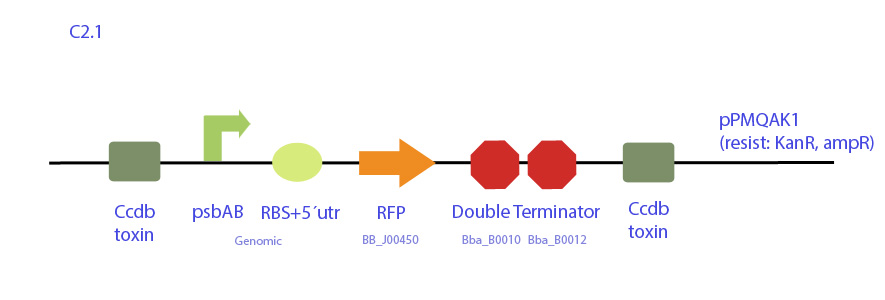
| - | + | [[File:UC_Chile-C2.1.jpg|center]] | |
| - | [[File: | + | <p>C2.1 |
| - | + | Construct designed from pPMQAK1 expression plasmid, which replicates both in E. coli and cyanobacteria and is biobrick-compatible. It codes for RFP under synechocystis psbAB promoter.</p> | |
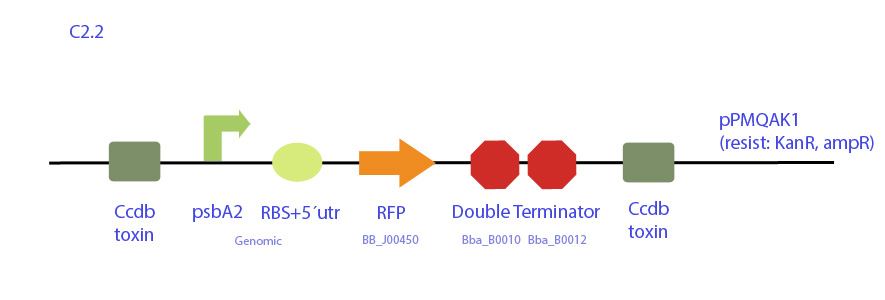
| - | <p | + | [[File:UC_Chile-C2.2.jpg|center]] |
| - | Construct designed from pPMQAK1 expression plasmid, which replicates both in E. coli and cyanobacteria and is biobrick-compatible. It codes for RFP under synechocystis psbAB promoter. | + | <p>C2.2 |
| - | + | Construct designed from pPMQAK1 expression plasmid, which replicates both in E. coli and cyanobacteria and is biobrick-compatible. It codes for RFP under synechocystis psaA2 promoter.</p> | |
| - | + | [[File:UC_Chile-C3.1.jpg|center]] | |
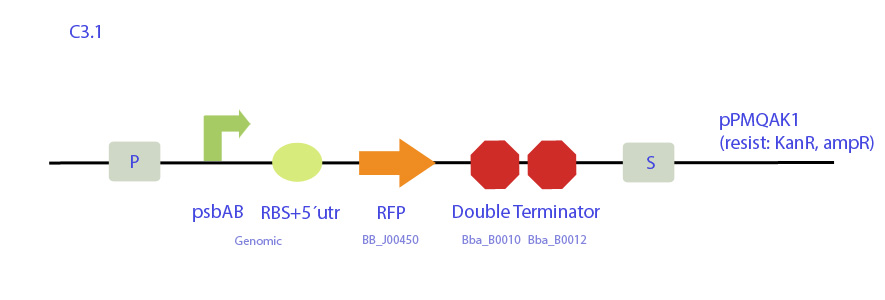
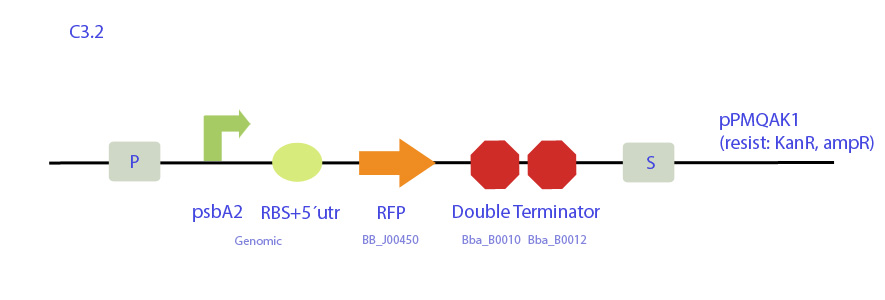
| - | [[File: | + | <p>C3 |
| - | + | Construct designed from pPMQAK1 expression plasmid, which replicates both in E. coli and cyanobacteria and is biobrick-compatible. It codes for RFP under synechocystis psbAB promoter and cuts out the whole Ccdb toxin gene.</p> | |
| - | <p | + | [[File:UC_Chile-C3.2.jpg|center]] |
| - | Construct designed from pPMQAK1 expression plasmid, which replicates both in E. coli and cyanobacteria and is biobrick-compatible. It codes for RFP under synechocystis psaA2 promoter. | + | |
| - | + | ||
| - | + | ||
| - | [[File: | + | |
| - | + | ||
| - | <p | + | |
| - | Construct designed from pPMQAK1 expression plasmid, which replicates both in E. coli and cyanobacteria and is biobrick-compatible. It codes for RFP under synechocystis psbAB promoter and cuts out the whole Ccdb toxin gene. | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | [[File: | + | |
[[File:c4.jpg]] | [[File:c4.jpg]] | ||
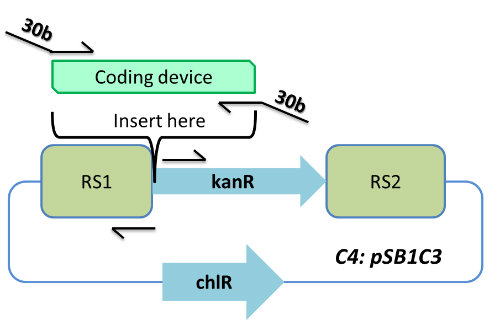
| - | <p | + | <p>C4<br> |
Expression plasmid for the expression of LuxA and LuxB genes under psbAB promoter. | Expression plasmid for the expression of LuxA and LuxB genes under psbAB promoter. | ||
| - | + | </p> | |
[[File:c5.jpg]] | [[File:c5.jpg]] | ||
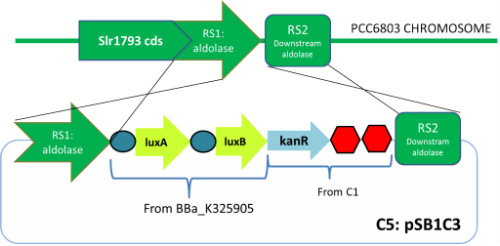
| - | <p | + | <p>C5<br> |
| - | Integrative plasmid designed to integrate LuxA and LuxB and a kanamycin resistance cassette downstream the 3' end of the transaldolase gene whose mRNA levels where seen to oscillate in a circadian manner peaking at hour 14, just after dusk. It is expect the same for LuxAB mRNA levels. | + | Integrative plasmid designed to integrate LuxA and LuxB and a kanamycin resistance cassette downstream the 3' end of the transaldolase gene whose mRNA levels where seen to oscillate in a circadian manner peaking at hour 14, just after dusk. It is expect the same for LuxAB mRNA levels.</p> |
| - | + | ||
<br> | <br> | ||
| - | <p | + | <p>C6<br> |
| - | Starting from construct C1, we designed an integrative plasmid that codes for LuxA and LuxB under the genomic transaldolase promoter (Pta). This time we decided to use the reverse complement of kanamycin resistance gene so both LuxAB and the resistance gene end at a double terminator sequence. You can check the problems we had with the standard kanR-double terminator (P1003+B0014) in our lab notebook. | + | Starting from construct C1, we designed an integrative plasmid that codes for LuxA and LuxB under the genomic transaldolase promoter (Pta). This time we decided to use the reverse complement of kanamycin resistance gene so both LuxAB and the resistance gene end at a double terminator sequence. You can check the problems we had with the standard kanR-double terminator (P1003+B0014) in our lab notebook.</p> |
| - | + | ||
<br> | <br> | ||
| - | <p | + | <p>C7.1<br> |
| - | Starting from psb1A3_intC integration plamid (BB_K390300) kindly provided to us by USU igem 2010, we designed a construct composed of LuxCDEG operon under synechocystis sigE promoter, whose mRNA abundance has been seen to oscillate in a circadian manner, with a peak at hour 8, before dusk. | + | Starting from psb1A3_intC integration plamid (BB_K390300) kindly provided to us by USU igem 2010, we designed a construct composed of LuxCDEG operon under synechocystis sigE promoter, whose mRNA abundance has been seen to oscillate in a circadian manner, with a peak at hour 8, before dusk.</p> |
| - | + | ||
<br> | <br> | ||
| - | <p | + | <p>C7.1<br> |
| - | Starting from psb1A3_intC integration plamid (BB_K390300) kindly provided to us by USU igem 2010, we designed a construct composed of LuxCDEG operon under synechocystis caa3 gene promoter, whose mRNA abundance has been seen to oscillate in a circadian manner, with a peak at hour 8, before dusk. | + | Starting from psb1A3_intC integration plamid (BB_K390300) kindly provided to us by USU igem 2010, we designed a construct composed of LuxCDEG operon under synechocystis caa3 gene promoter, whose mRNA abundance has been seen to oscillate in a circadian manner, with a peak at hour 8, before dusk.</p> |
| - | + | ||
<br> | <br> | ||
| + | {{UC_Chilefooter}} | ||
Latest revision as of 23:50, 21 September 2012
 "
"