|
|
| (4 intermediate revisions not shown) |
| Line 16: |
Line 16: |
| | <br /> | | <br /> |
| | [[#Temperature| Temperature]] | | [[#Temperature| Temperature]] |
| | + | |
| | + | |
| | | | |
| | <br /> | | <br /> |
| Line 62: |
Line 64: |
| | | | |
| | | | |
| - | The results confirm our hypothesis: measured luminiscence output is growth-state dependent, low at the early lag phase, peaking at early log phase and decaying at late stationary phase. | + | The results confirm our hypothesis: measured luminescence output is growth-state dependent, low at the early lag phase, peaking at early log phase and decaying at late stationary phase. |
| | | | |
| | </div> | | </div> |
| Line 94: |
Line 96: |
| | | | |
| | Lux genes were originally cloned from <i>Vibrio fischerii</i>, a free living bacteria found in tropical sea water that also inhabits the poikilotherm mollusc <i>Euprymna scolopes</i> body in a symbiotic relationship, therefore we expect the Lux genes performance to be functional in the range of temperatures found in that environmental context. | | Lux genes were originally cloned from <i>Vibrio fischerii</i>, a free living bacteria found in tropical sea water that also inhabits the poikilotherm mollusc <i>Euprymna scolopes</i> body in a symbiotic relationship, therefore we expect the Lux genes performance to be functional in the range of temperatures found in that environmental context. |
| - | It has been shown that the optimal temperature conditions for bioluminescence in <i>V. harveyi</i> range from 20-26°C [[#5|5]] and the LuxBrick page states that luminescence is diminished in temperatures above 30°C. We aimed to characterize the luminiscence output of the LuxBrick in a set of temperature conditions used in <i>E.coli</i> cultivation. | + | It has been shown that the optimal temperature conditions for bioluminescence in <i>V. harveyi</i> range from 20-26°C [[#5|5]] and the LuxBrick page states that luminescence is diminished in temperatures above 30°C. We aimed to characterize the luminescence output of the LuxBrick in a set of temperature conditions used in <i>E.coli</i> cultivation. |
| | </div> | | </div> |
| | <br> | | <br> |
| | + | <h1>N-decanal Effect Characterization - (BBa_K325909 and BBa_K325905)</h1> |
| | + | </div> |
| | + | We decided to make a luciferase bioluminescence assay to put to test wether [http://partsregistry.org/Part:BBa_K743003 Pta promoter] is effectively driving LuxAB genes expression in [http://partsregistry.org/Part:BBa_K743014 BBa_K743014] and [http://partsregistry.org/Part:BBa_K743015 BBa_K743015] transformed into Synechocystis. |
| | + | At the same time, we made the same assay to test if the [http://partsregistry.org/Part:BBa_K325905 Bacterial Lux reporter CDEG pλ AB] from wich we cloned the LuxAB genes produces light emition in E. coli cells under different n-decanal concentrations. |
| | + | |
| | + | '''Results''' |
| | + | |
| | + | <html><img src="http://partsregistry.org/wiki/images/d/d6/Potopotopoto.jpg" width="500"></html> |
| | + | |
| | + | <html><img src="http://partsregistry.org/wiki/images/8/87/Cacacacax56t.jpg" width="500"></html> |
| | + | |
| | + | '''Analysis''' |
| | + | Results of our measurement showed production and expression of Luciferase only for LuxBrick. This BioBrick has a basal luminescence yield which is augmented in the presence of decanal and dodecanal. |
| | + | The results suggest that the limiting factor in the Luciferase reaction is the substrate production and not the catalytic activity of the enzyme, as is documented in literature. |
| | + | |
| | + | For the rest of the constructs measurements show total absence of luminescence for the whole range of aldehyde concentrations suggesting that luciferases are not being expressed. We believe this is caused by a failure in the [http://partsregistry.org/Part:BBa_K743003 Pta promoter] region which was taken 200 bp upstream from the original gene. We think that maybe the actual promoter could have been interrupted therefore it will be amplified again from chassis genome taking a much larger sequence upstream the atg start codon. |
| | + | |
| | + | Part [http://partsregistry.org/Part:BBa_K325905 K325905] is a Bacterial Lux reporter |
| | + | CDEG pλ AB that contains a luciferase gene(from Xenorhabdus luminescens) under plambda promoter. The presence of decanal or dodecanal should trigger luminescence production. However, luminescence levels are practically zero, which could be attributed to an error in the LuxAB gene or in the promoter. |
| | + | As [http://partsregistry.org/Part:BBa_K743014 BBa_K743014] was built from [http://partsregistry.org/Part:BBa_K325905 K325905] it is possible that our construct inherited the problems that make this biobrick fail |
| | + | |
| | <h1>References</h1> | | <h1>References</h1> |
| | <div id="1"> | | <div id="1"> |
| Line 117: |
Line 140: |
| | </div> | | </div> |
| | (5) Scheerer S, Gomez F, Lloyd D. Bioluminescence of Vibrio fischeri in continuous culture: optimal conditions for stability and intensity of photoemission. Journal of Microbiol Methods. 2006 Nov;67(2):321-9. Epub 2006 Jun 5. | | (5) Scheerer S, Gomez F, Lloyd D. Bioluminescence of Vibrio fischeri in continuous culture: optimal conditions for stability and intensity of photoemission. Journal of Microbiol Methods. 2006 Nov;67(2):321-9. Epub 2006 Jun 5. |
| | + | |
| | | | |
| | | | |
| Line 122: |
Line 146: |
| | | | |
| | <html> | | <html> |
| - | <a href="https://2012.igem.org/Team:UC_Chile/Results/Vs"><img src="https://static.igem.org/mediawiki/2012/a/ab/UC_Chile-Continue_button.jpg" align="right"> | + | <a href="https://2012.igem.org/Team:UC_Chile/Protocols"><img src="https://static.igem.org/mediawiki/2012/a/ab/UC_Chile-Continue_button.jpg" align="right"> |
| | </html> | | </html> |
| | | | |
| | {{UC_Chilefooter}} | | {{UC_Chilefooter}} |

LuxBrick Characterization - (BBa_K325909)
As a part of our project, we set out to investigate the conditions which affect the output of the LuxBrick. As we are using various elements from this Biobrick, we believe that is cornersome to know how we may increase its output by modifying the conditions that affect its behaviour.
We set out to understand diverse factors that could be influencing the total bioluminescence yield of the brick. Some of them have direct relation with specific of the aspects of our project. These factors are:
Glucose concentration
Growth state at time of induction
Cell mass
Temperature
Here you'll find detailed the characterization methods we used in this experiment.
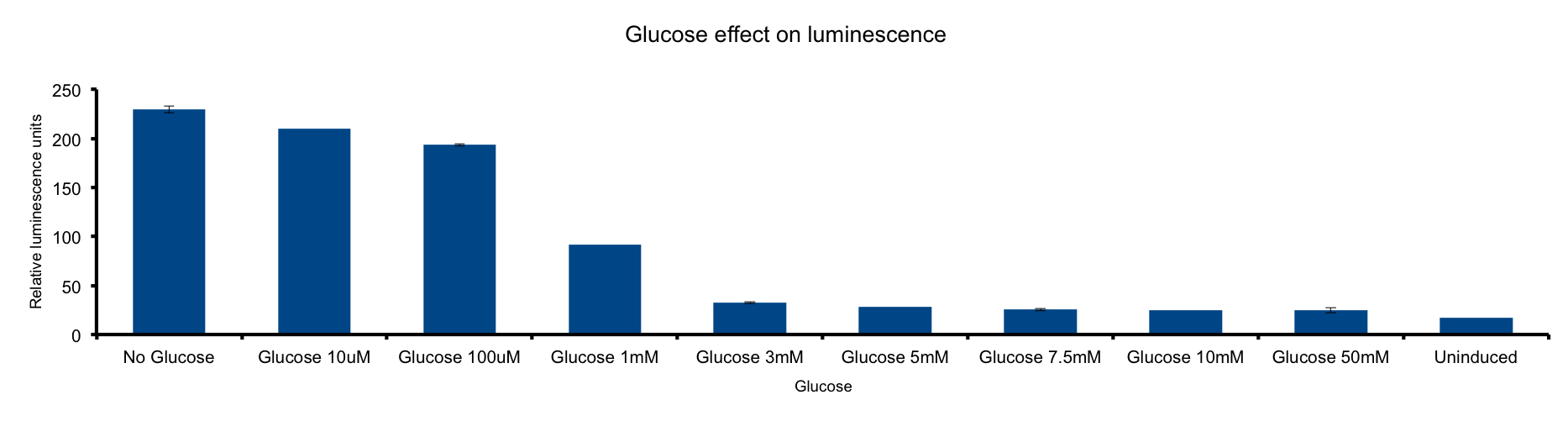
Glucose concentration in media
Glucose is a molecule central for the energy metabolism throughout living organisms and its the main energy source for E.coli.
As reduced carbon-hydrogen bonds in glucose are oxidized through glicolisis and TCA cycle, reduced equivalents are stored in NADPH and FADH2. The majority of these electrons finally react with molecular oxygen to form water at the end of the electron transport chain (ETC), a reaction catalized by citochrome c oxydase.
This metabolic pathway is linked to the Lux light emission in two ways: the luciferase reaction consumes reducing power and on the other hand it can be considered respiration as it consumes oxygen 1.
Our results clearly show a strong inhibition of light-emittion down to uninduced levels at glucose concentrations higher than 3mM. Our hypothesis is that ETC respiration competes for oxygen with luciferase respiration; if glucose is added to the medium, the reduction of oxygen to water would be enhanced making oxygen less available to the luciferase reaction.
The fact that LuxAB light-emittion is augmented by cytochrome c oxydase inhibition supports this idea 2.
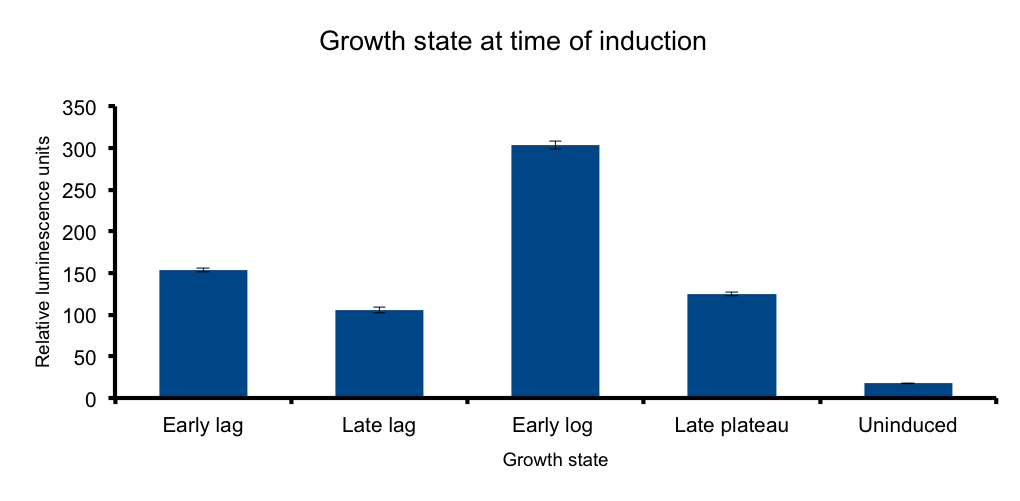
Growth state at-time-of-induction
It has been know for some time that general bacterial metabolism is determined by its growth state 3. Growth
phase determines available resources and stress derived restrictions. Bacteria can dramatically adjust their metabolism to adapt to these changing conditions.
At the transcriptional level, sigma subunits present at different growth states regulate gene expression throughout the whole genome 4.
We hypothesized that the heterologous expression of the Lux operon will also be subject to the regulation exerted by the metabolic state of the bacteria.
The results confirm our hypothesis: measured luminescence output is growth-state dependent, low at the early lag phase, peaking at early log phase and decaying at late stationary phase.
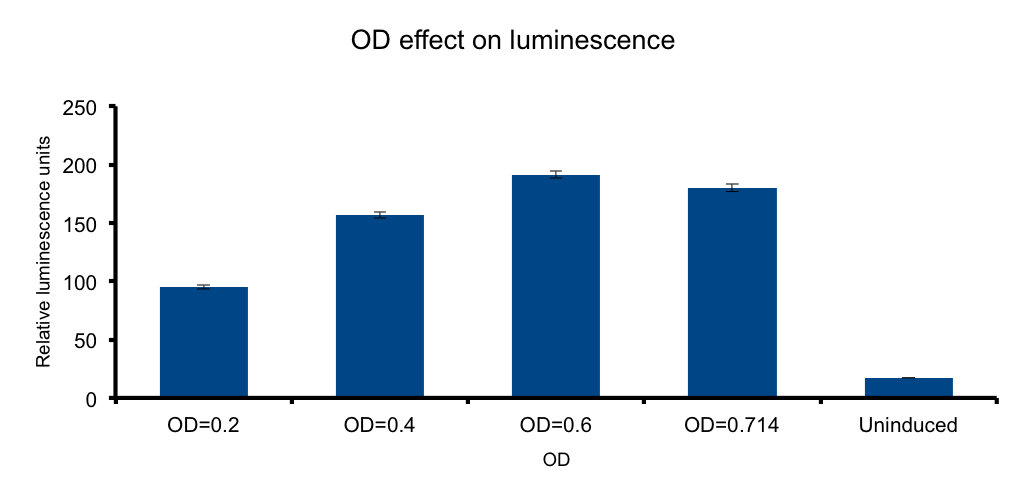
Cell mass (Optical Density)
If cells diluted in a liquid media behave as light-emitting entities, light transmittion to the exterior will be a function of cell density of the media.
We wanted to know if there is an optimal density, under which the emittion of photons by the total number of bacteria versus the absorbance of the emitted photons by the same bacteria in the media, peak. In such a scenario any number of bacteria in excess beyond that point will not contribute to increase light output, and the system will lose efficiency.
Our measurements showed that such a hypothesis is plausible, where a clear increase of light output is seen towards higher ODs. However, the data shows that the output reaches a maximum at OD 0.6, but by the closeness of the signal of the OD 0.714 and the scarceness of the experimental point measurements, we cannot conclude that at OD 0.6 the maxima is reached. Further OD points ought to be tested to reach such conclusions.
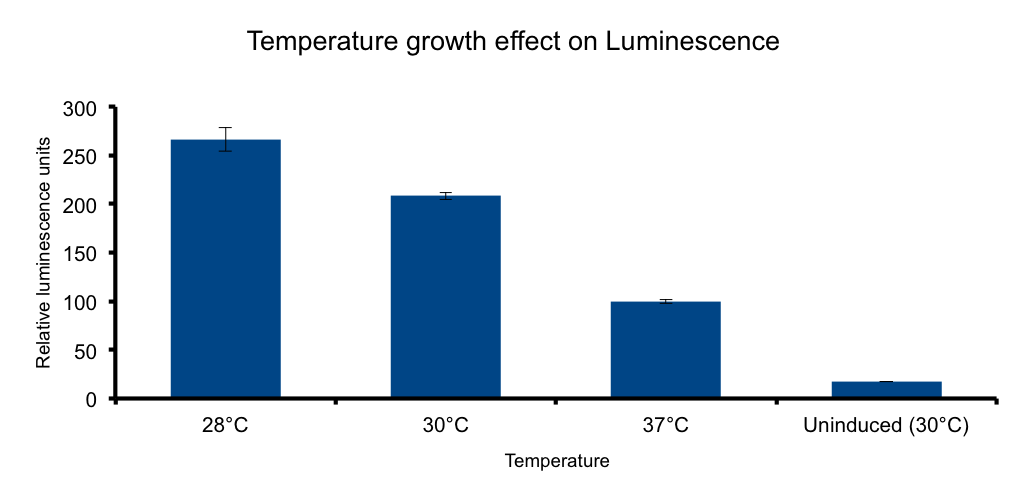
Growth temperature
Lux genes were originally cloned from Vibrio fischerii, a free living bacteria found in tropical sea water that also inhabits the poikilotherm mollusc Euprymna scolopes body in a symbiotic relationship, therefore we expect the Lux genes performance to be functional in the range of temperatures found in that environmental context.
It has been shown that the optimal temperature conditions for bioluminescence in V. harveyi range from 20-26°C 5 and the LuxBrick page states that luminescence is diminished in temperatures above 30°C. We aimed to characterize the luminescence output of the LuxBrick in a set of temperature conditions used in E.coli cultivation.
N-decanal Effect Characterization - (BBa_K325909 and BBa_K325905)
</div>
We decided to make a luciferase bioluminescence assay to put to test wether Pta promoter is effectively driving LuxAB genes expression in BBa_K743014 and BBa_K743015 transformed into Synechocystis.
At the same time, we made the same assay to test if the Bacterial Lux reporter CDEG pλ AB from wich we cloned the LuxAB genes produces light emition in E. coli cells under different n-decanal concentrations.
Results


Analysis
Results of our measurement showed production and expression of Luciferase only for LuxBrick. This BioBrick has a basal luminescence yield which is augmented in the presence of decanal and dodecanal.
The results suggest that the limiting factor in the Luciferase reaction is the substrate production and not the catalytic activity of the enzyme, as is documented in literature.
For the rest of the constructs measurements show total absence of luminescence for the whole range of aldehyde concentrations suggesting that luciferases are not being expressed. We believe this is caused by a failure in the Pta promoter region which was taken 200 bp upstream from the original gene. We think that maybe the actual promoter could have been interrupted therefore it will be amplified again from chassis genome taking a much larger sequence upstream the atg start codon.
Part K325905 is a Bacterial Lux reporter
CDEG pλ AB that contains a luciferase gene(from Xenorhabdus luminescens) under plambda promoter. The presence of decanal or dodecanal should trigger luminescence production. However, luminescence levels are practically zero, which could be attributed to an error in the LuxAB gene or in the promoter.
As BBa_K743014 was built from K325905 it is possible that our construct inherited the problems that make this biobrick fail
References
(1) Makemson, J C. Luciferase-dependent oxygen consumption by bioluminescent vibrios.
J. Bacteriol. February 1986 vol. 165 no. 2 461-466
(2) Ulitzur, S. A. Reinhertz and J. W. Hastings.Factors affecting the cellular expression of bacterial luciferase
Archives of Microbiology. Volume 129, Number 1 (1981), 67-71
(3) Rahman, Mahbuba; Rubayet, Mohammad; Hasan and Kazuyuki Shimizu.Growth phase-dependent changes in the expression of global regulatory genes and associated metabolic pathways in Escherichia coli.Biotechnology letters.
Volume 30, Number 5 (2008)
</div>
(4) Chang, Dong-Eun; Darren J. Smalley; Tyrrell Conway. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Molecular Microbiology. Volume 45, Issue 2, pages 289–306, July 2002.
(5) Scheerer S, Gomez F, Lloyd D. Bioluminescence of Vibrio fischeri in continuous culture: optimal conditions for stability and intensity of photoemission. Journal of Microbiol Methods. 2006 Nov;67(2):321-9. Epub 2006 Jun 5.

 "
"