Team:UC Chile/Results/Growthcurve
From 2012.igem.org
(Difference between revisions)
| Line 12: | Line 12: | ||
Methods for the growth curve characterization of Synechocystis PCC 6803 over [[Team:UC_Chile2/Protocols#SyneGrowth | here]]. | Methods for the growth curve characterization of Synechocystis PCC 6803 over [[Team:UC_Chile2/Protocols#SyneGrowth | here]]. | ||
| + | |||
| + | <font size="3">Results</font> | ||
| + | |||
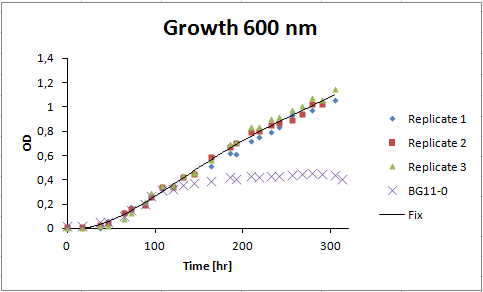
| + | Model for Synechocystis growth at OD 600 nm and 730 nm | ||
| + | |||
| + | After analyzing the experimental data, a polinomial growth model was obtained by function curve fitting. We used Excel's Solver tool for data processing | ||
| + | |||
| + | Model at 600 nm : eq | ||
<html><img src="" align="center"></html> | <html><img src="" align="center"></html> | ||
[[File: Figure for the growth curve.jpg| 500px |center]] | [[File: Figure for the growth curve.jpg| 500px |center]] | ||
| + | |||
| + | Model at 730 nm : eq | ||
| + | |||
| + | <html><img src="" align="center"></html> | ||
| + | |||
| + | Exponential phase growth curve | ||
| + | |||
| + | As we wanted to see the behaviour of cells in exponential phase, we analysed the part of the prior curves which best fitted the exponential function. We considered a first order kinetics growth. | ||
| + | |||
| + | The expression that governs the exponential growth given by the expression: | ||
| + | |||
| + | Adjust of mu parameter was obtained using Solver's algorithm of least squares. | ||
| + | |||
| + | eq | ||
| + | |||
| + | Exponential growth at 600 nm | ||
| + | |||
| + | Exponential growth at 730 nm | ||
| + | |||
| + | The exponential model allows us to express duplication time as the independent variable by equation rearragement. | ||
| + | |||
| + | |||
| + | Relation of measurements at 600 nm and 730 nm | ||
| + | |||
| + | In other data analysis, we wanted to check of we could move from one OD to the other. This is, that we can predict OD at 730 nm with 600 nm data. As it happens, this is possible and the linear model between measurements is given by | ||
| + | |||
| + | eq | ||
| + | |||
| + | |||
| + | |||
{{UC_Chilefooter}} | {{UC_Chilefooter}} | ||
Revision as of 18:12, 26 September 2012
 "
"