Team:Tuebingen/ProjectImplementation
From 2012.igem.org
Revision as of 09:27, 24 September 2012 by Jakobmatthes (Talk | contribs)
Implementation
Contents |
The following plasmid constructs are needed to implement the measurement system. Using two receptors, two signalling systems and two reporter genes the implementation contains redundancy to select for a sensitive system.
Plasmid 1
Task: Expression of membrane receptor.
| backbone pRS# | part #1 | part #2 | part #3 |
|---|---|---|---|
| 313 | Padh1 | mPR Danio rerio | Tadh1 |
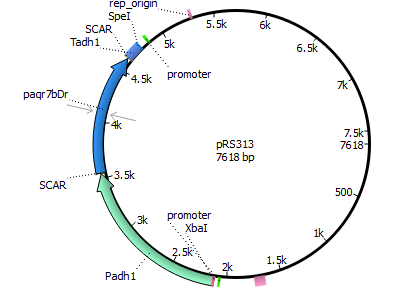
| |||
| 313 | Padh1 | mPR Xenopus laevis | Tadh1 |
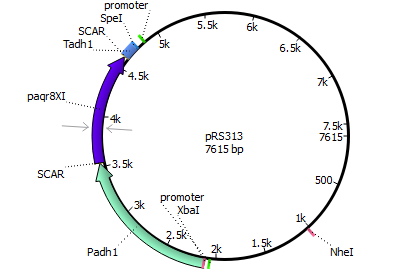
| |||
Plasmid 2
Task: Signalling, inhibitor controlled by membrane receptor through Pfet3.
| backbone pRS# | part #1 | part #2 | part #3 |
|---|---|---|---|
| 315 | Pfet3 | rox1 | Tadh1 |
targets Plasmid 3 with Panb1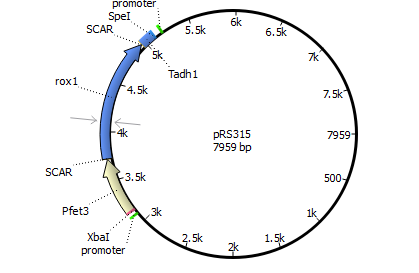
| |||
| 315 | Pfet3 | mig1 | Tadh1 |
targets Plasmid 3 with Psuc2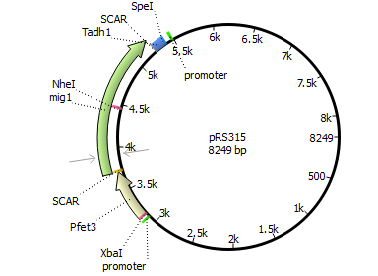
| |||
Plasmid 3
Task: Expression of reporter gene, controlled by inhibitor (plasmid 2).
| backbone pRS# | part #1 | part #2 | part #3 |
|---|---|---|---|
| 316 | Panb1 | luciferase | Tadh1 |
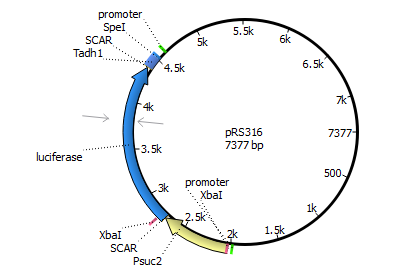
| |||
| 316 | Panb1 | lacZ | Tadh1 |
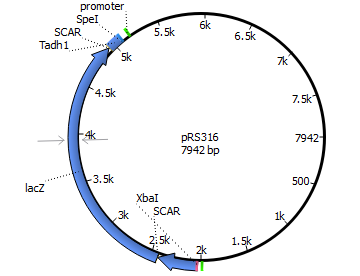
| |||
| 316 | Psuc2 | luciferase | Tadh1 |

| |||
| 316 | Psuc2 | lacZ | Tadh1 |
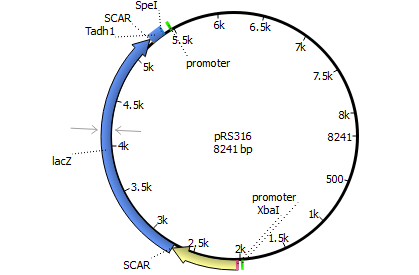
| |||
Measurement
The measurement itself should be limited to an optical one. With this idea in mind, we decided for reporter genes like lacZ and luciferase, because these produce signals which can simply be quantified by optical measurement methods.
 "
"