Team:Tianjin/Modeling/Regulation
From 2012.igem.org

Contents |
Background
Metabolic Regulation
Cell metabolism is the foundation of cell growth, secretion, differentiation, etc. as well as the core process of the modern fermentation technology that can make large impact on the quality and output of product.
However the modern metabolic regulation of strains has a large amount of areas for improvement. Take the most common E. coli as example. At present, most of the regulation means is to use a single promoter to construct specific operon gene cluster to achieve the regulation with the addition of certain inducers induction of a specific protein. But this induced expression is typically unidirectional, irreversible and it needs to build many complex operons’ gene structure to construct multiple logical regulation, this will produce a plenty of limitation.
If we add our O-Key into the expression system, a novel regulation means will exist on the level of translation. Meanwhile, we can combine O-Key with the traditional transcription regulation to create logic gate regulation structure like “AND” gate. The logic gate working with normal close and normal open promoter can constitute multiple “AND/OR/NOR” logic gate control system which has more simple structure compared to traditional regulation.
The Difficulty of Large Fragments Assembly
The general digestion connection will leave scar and will be limited by the specific cleavage sequences.
Long PCR fragment will suffer the decline of success rate and distortion, etc.
However, the construction of some large fragments cannot be avoided, so the development of a low-cost, simple operation, good fidelity, a little limiting factor large DNA fragment assembly method is particularly important.
Yeast Assembler
History
Yeast Assembler is based on in vivo homologous recombination in yeast. As for its high efficiency and ease to work with, in vivo homologous recombination in yeast has been widely used for gene cloning, plasmid construction and library creation. In the early of 2008, Zengyi Shao from University of lllinois at Urbana-Champaign, Urbana, used such a method to construct biochemical pathways. Such a method, for its high efficiency in assembling multiple genes, received great popularity since its appearance.
Principles
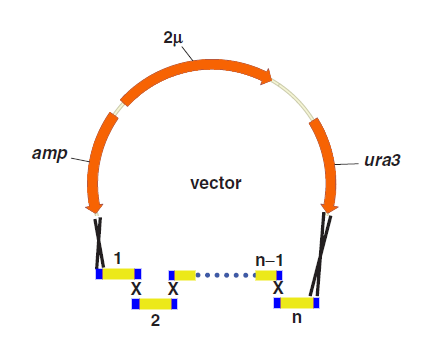
One step assembly into a vector.
When parts are transformed all parts into Yeast, homologous recombination occurs at the site “x”, and then all little parts are integrated into a vector.
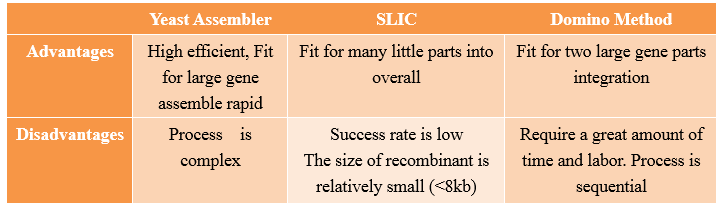
Advantages and Disadvantages
Compared with other methods,the “Yeast assembler” are more efficient and useful for large gene assemble.
 "
"