Team:Tianjin/Data
From 2012.igem.org

BioBrick
Favorite Tianjin 2012 iGEM Team Parts
| Name | Type | Description | Designer | Length | ||
|---|---|---|---|---|---|---|
| 1 | W | BBa_K821001 | Reporter | RFP Reporter with o-RBS | Qinfeng Wu, Bin Jia | 1059 |
| 2 | W | BBa_K821002 | Coding | Orthogonal 16S DNA sequence from rrnB | Qinfeng Wu, Bin Jia | 1894 |
| 3 | W | BBa_K821012 | Generator | PBad(promoter)+orthogonal 16S RNA | Jinlai Zhang, Dongchang Qin | 3226 |
Tianjin 2012 iGEM Team Parts Sandbox
| Name | Type | Description | Designer | Length | ||
|---|---|---|---|---|---|---|
| 1 | W | BBa_K821003 | Coding | 16S DNA sequence from rrnB | Jinlai Zhang, Dongchang Qin | 1894 |
| 2 | W | BBa_K821004 | Regulatory | PBad Promoter | Jinlai Zhang, Dongchang Qin | 1324 |
| 3 | W | BBa_K821005 | Reporter | CmR label from pKD3 | Qinfeng Wu, Bin Jia | 1014 |
| 4 | W | BBa_K821006 | Project | KanR label from pKD3 | Jinlai Zhang, Dongchang Qin | 1304 |
| 5 | W | BBa_K821007 | Composite | PL(promoter)+orthogonal 16S RNA | Qinfeng Wu, Bin Jia | 1957 |
| 6 | W | BBa_K821008 | Composite | PLtet+orthogonal 16S RNA | Jinlai Zhang, Dongchang Qin | 1956 |
| 7 | W | BBa_K821009 | RBS | Orthogonal RBS 1 | Qinfeng Wu, Bin Jia | 6 |
| 8 | W | BBa_K821010 | Composite | PLacI+Native RBS+RFP+Native RBS+GFP+Terminator | Qinfeng Wu, Bin Jia | 1833 |
| 9 | W | BBa_K821003 | Composite | PLacI+Orthogonal RBS+RFP+Native RBS+GFP+Terminator | Jinlai Zhang, Dongchang Qin | 1827 |
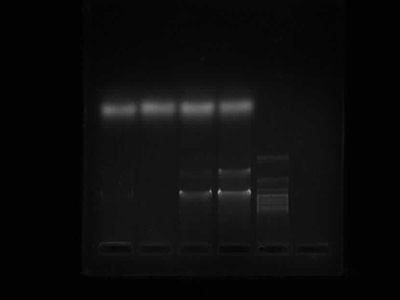
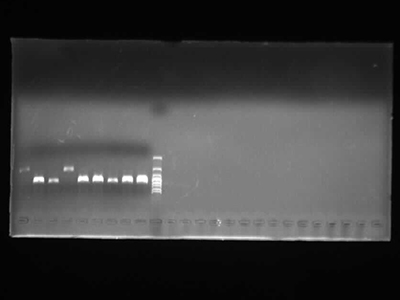
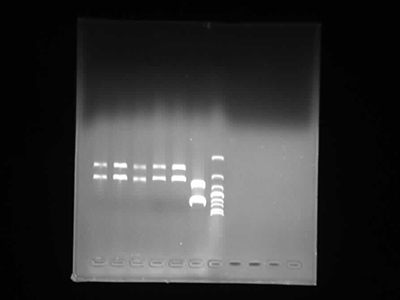
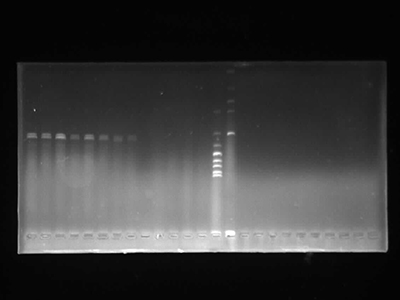
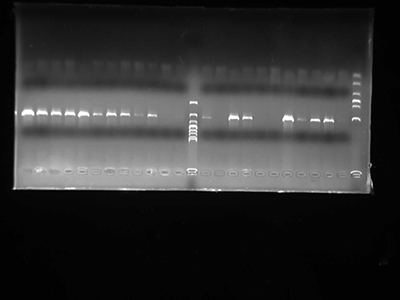
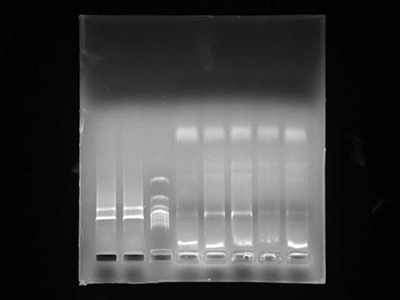
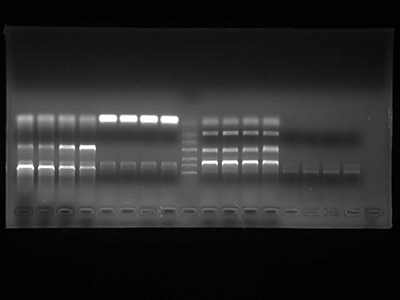
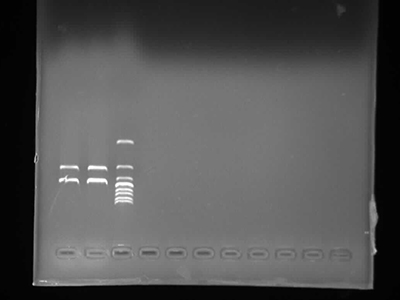
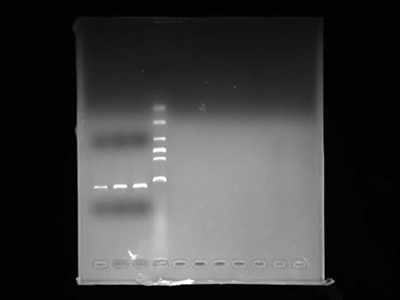
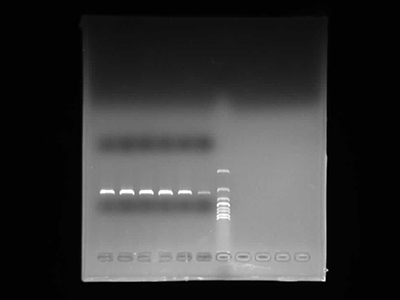
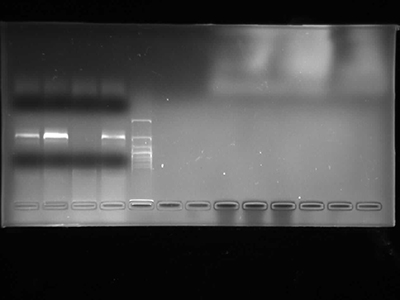
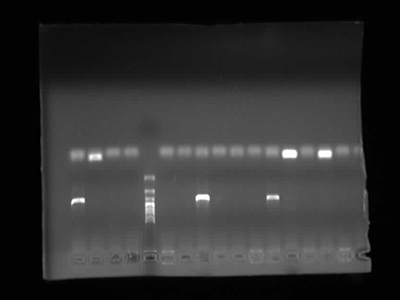
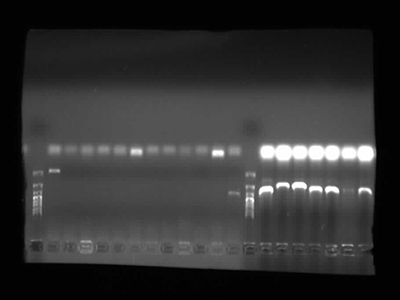
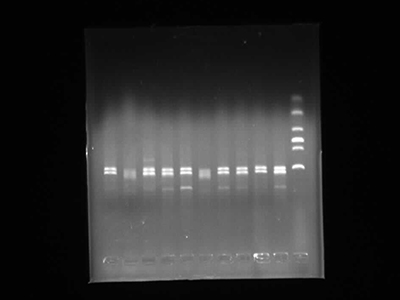
Agarose Gel Electrophoresis
The Difficulty of Large Fragments Assembly
The general digestion connection will leave scar and will be limited by the specific cleavage sequences.
Long PCR fragment will suffer the decline of success rate and distortion, etc.
However, the construction of some large fragments cannot be avoided, so the development of a low-cost, simple operation, good fidelity, a little limiting factor large DNA fragment assembly method is particularly important.
Yeast Assembler
History
Yeast Assembler is based on in vivo homologous recombination in yeast. As for its high efficiency and ease to work with, in vivo homologous recombination in yeast has been widely used for gene cloning, plasmid construction and library creation. In the early of 2008, Zengyi Shao from University of lllinois at Urbana-Champaign, Urbana, used such a method to construct biochemical pathways. Such a method, for its high efficiency in assembling multiple genes, received great popularity since its appearance.
Principles
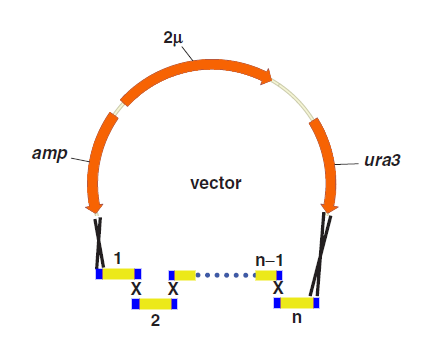
One step assembly into a vector.
When parts are transformed all parts into Yeast, homologous recombination occurs at the site “x”, and then all little parts are integrated into a vector.
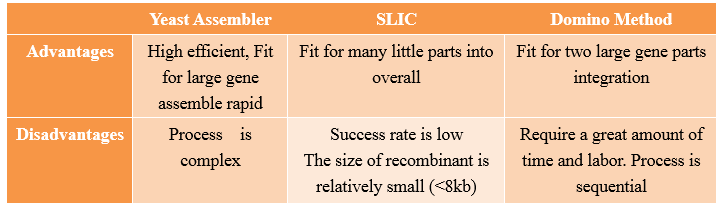
Advantages and Disadvantages
Compared with other methods,the “Yeast assembler” are more efficient and useful for large gene assemble.
Completely Synthesizing the Genome of Mycoplasma Genitalium using Yeast Assembler
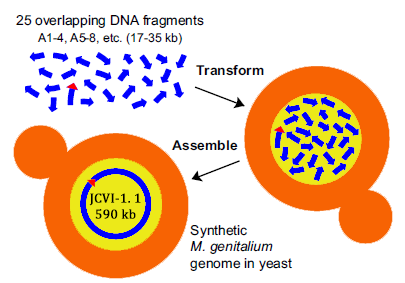
In 2008, Gibson from the J. Craig Venter Institute, published an article “one-step assembly in Yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome”. In the article, the author transformed 25 overlapping DNA fragments into Yeast, homologous recombination occurs, and then the whole genome is synthesized.
Construction of a synthetic M. genitalium genome in yeast. Yeast cells were transformed with 25 different overlapping A-series DNA segments (blue arrows; ~17 kb to35 kb each) composing the M. genitalium genome. To assemble these into a complete genome, a single yeast cell (tan) must take up at least one representative of the 25 different DNA fragments and incorporate them in the nucleus (yellow), where homologous recombination occurs. This assembled genome, called JCVI-1.1, is 590,011 bp, including the vector sequence (red triangle) shown internal to A86 – 89.
 "
"