Team:Technion/Modeling
From 2012.igem.org

Most of our modelling efforts were done for the riboswitch system. The modelling helped us in the debugging process and in some planing stages.
Contents |
Back of the envelope calculations
In order to assess the ration of theophylline molecules to riboswitch containing RNA molecules, some "back of the envelope" calculations were done. These calculations were under the following assumptions:
- 1 nm of theophylline = 1 molecule per cell => 1mM = 106 molecules per cell. This assumption is based on the BioNumbers database {1, 2}.
- The RNA half life in a cell is approximately 5 minutes {2}. Therefore, the degradation rate is ~0.14(1/min).
- The RNA synthesis rate is ~1(molecule/min) considering the mRNA length for the RS+mCherry construct.
Assuming a steady state, the amount of RNA molecules in a cell would be the synthesis rate divided by the degradation rate. This would produce ~7 RNA molecules per DNA template.
Next, the number of DNA templates was approximated using [http://endmemo.com/bio/dnacopynum.php this calculator]. The amount of DNA from pSB1C3 minipreps was ~5000ng for a plasmid+insert size of 2888bp. The amount of DNA from pSB3C5 minipreps was ~12500ng for a plasmid+insert size of 3556bp. Using the calculator, the copy number of the plasmids was ~160 and ~325 for pSB1C3 and pSB3C5 respectively.
Assuming that not all the copies of the plasmid are transcribed simultaneously (therefore dividing the copy number by 3), the number of RNA molecules per cell was 366 and 745 for pSB1C3 and pSB3C5 respectively. Therefore, the ratio between the theophylline molecules and the mRNA molecules at 1mM theophylline is ~2730 and ~1340 for pSB1C3 and pSB3C5 respectively.
This means that there are a lot of theophylline molecules in the cells to interact with the riboswitch. The level of mRNA molecules that are actually translated would depend on the riboswitch-theophylline Kd.
Secondary structures
In order to understand the behavior of our RS+mCherry construct, we examined the secondary structure of our system and compared it to systems that have been shown to work with the riboswitch. The secondary structures were calculated using [http://www.nupack.org/home/references NUPACK]{3}.
The comparison between different secondary structures is presented in Figure 1.
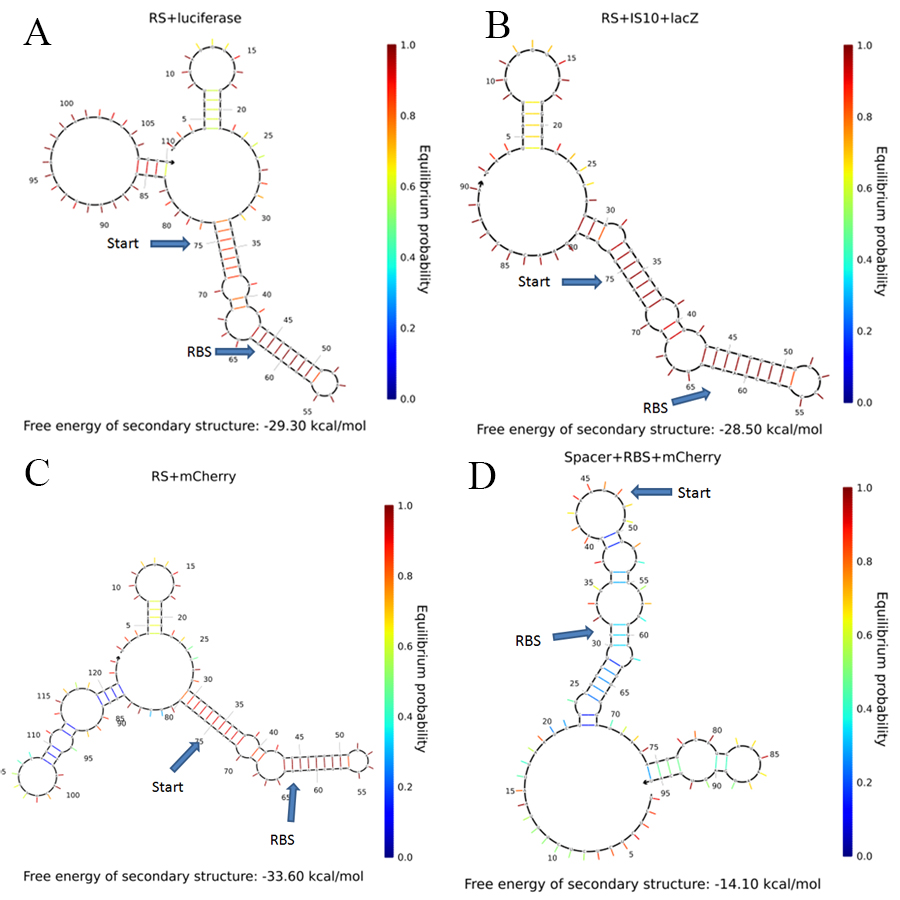
It can be seen that there are only minor differences in the secondary structure as predicted by the model. However, this does not mean that there isn't some other effect of the mRNA folding on the riboswitch function. Moreover, for the positive control (D) it can be seen that all the interactions are at low probability. Therefore, this RBS should be free for interaction with the ribosome most of the time.
RBS strength calculations
To further investigate the behavior of the RS+mCherry construct and its positive control (RBS+mCherry) we submitted the mRNA sequences to the [https://salis.psu.edu/software/ Salis lab RBS calculator]. Table 1 summarizes the translation initiation rates at the expected start codon for each construct.
| Construct | Translation initiation rate |
RS+luciferase |
319 |
RS+IS10+lacZ |
139 |
RS+mCherry |
56.7 |
RBS+mCherry |
199 |
It can be seen that the model gives low translation initiation rates for all the constructs. For the riboswitch constructs this is due to the secondary structures and the fact the calculation is done in the absence of theophylline. For the RBS+mCherry construct it is visible that the translation initiation rate is ~3.5 higher. Therefore, according to the model, the fluorescence from this construct should be higher than the one measured for the riboswitch in the absence of theophylline. However, this is in contrast to some of our experimental results.
Conclusion
We tried to use computational analysis tools such as [http://www.nupack.org/home/references NUPACK] and the [https://salis.psu.edu/software/ Salis lab RBS calculator] to explain and predict our experimental results. However, we saw that these tools did not provide us with a clear explanation. Therefore, we conclude that these tools should be used with caution and are good for general estimations.
References
- Suess B., et al. 2004. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Research 32(4): 1610-1614.
- Milo R., et al. 2009. BioNumbers--the database of key numbers in molecular and cell biology. Nucleic Acids Research 38(Database): D750-D753.
- Zadeh J. N., et al. 2011. NUPACK: Analysis and design of nucleic acid systems. Journal of Computational Chemistry 32(1): 170-173.
- Lynch S. A., Gallivan J. P. 2009. A flow cytometry-based screen for synthetic riboswitches. Nucleic Acids Research 37(1): 184-192.
 "
"