Team:TU Munich/Project/Constitutive Promoter
From 2012.igem.org



Contents |
Constitutive Promoters
Background and principles
Georg
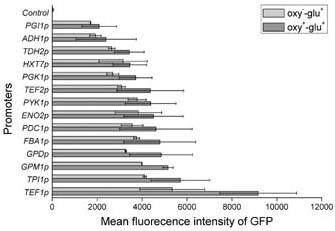
Jie Sun et al. describes 14 constitutive promoters from Saccharomyces cerevisiae, and its reaction at different culture conditions, oxy(-)/glu(+),oxy(+)/glu(+). 2 Where described as higher strength promoters TEF1p, TPI1p, and 7 as medium strenth pr. GPM1p, GPDp (TDH3p), FBA1p, PDC1p, ENO2p, PYK1p, and TEF2p. With exception to the strongest TEF1p all exhibit nearly the same expression values with varying oxygen conditions. TEF1p was also tested for small glucose concentrations showed 100% expression levels with oxygen limitation and 70 % without oxygenlimitation after 80 hours of cell cultivation.
A.Blount et al. states that repeated use of the same biological parts is not possible since the cells recombine long stretches of homologous DNA
Siavash Partow et al. compares several constitutive Promoters during varying Glucose concentrations Media:Tum12Promoterstärken.pdf, TEF1, HXT7,PGK1 are shown to be the most suitable ones. They also show the fusion of two promoters for bidirectional control of two genes namely TEF1 and PGK1 with comparable activity in both states Media:Tum12Promoterfusion.pdf, two constructs were successfully used pSP-G1-Vector and pSP-G2.
.
Media:Tum12Fusionspromoter_Vector.pdf
Elke Nevoigt et al. generated a library of TEF1 promotermutants for finetuning of Expression Media:Tum12 TEF-Promoter-mutants.pdf
Idea
General remarks and issues
TEF1p guarantues relative strong expression values under varying conditions as it is the case for brewing.
Biobricks and sequences
TEF1 sequence : from Promoter Database for Sacharomyces cerevisiae SCPD :(http://rulai.cshl.edu/cgi-bin/SCPD/getgenelist)
RAP1
GCCGTACCACTTCAAAACACCCAAGCACAGCATACTAAATTTCCCCTCTT -301
TCTTCCTCTAGGGTGTCGTTAATTACCCGTACTAAAGGTTTGGAAAAGAA -251
AAAAGAGACCGCCTCGTTTCTTTTTCTTCGTCGAAAAAGGCAATAAAAAT -201
TTTTATCACGTTTCTTTTTCTTGAAAATTTTTTTTTTTGATTTTTTTCTC -151
TTTCGATGACCTCCCATTGATATTTAAGTTAATAAACGGTCTTCAATTTC -101
TCAAGTTTCAGTTTCATTTTTCTTGTTCTATTACAACTTTTTTTACTTCT -51
TGCTCATTAGAAAGAAAGCATAGCAATCTAATCTAAGTTTTAATTACAAA
TPI1 Promotorsequenz
CTACTTATTCCCTTCGAGATTATATCTAGGAACCCATCAGGTTGGTGGAA -401
GCR1 RAP1
GATTACCCGTTCTAAGACTTTTCAGCTTCCTCTATTGATGTTACACCTGG -351
RAP1 GCR1
ACACCCCTTTTCTGGCATCCAGTTTTTAATCTTCAGTGGCATGTGAGATT -301
CTCCGAAATTAATTAAAGCAATCACACAATTCTCTCGGATACCACCTCGG -251
TTGAAACTGACAGGTGGTTTGTTACGCATGCTAATGCAAAGGAGCCTATA -201
TACCTTTGGCTCGGCTGCTGTAACAGGGAATATAAAGGGCAGCATAATTT -151
AGGAGTTTAGTGAACTTGCAACATTTACTATTTTCCCTTCTTACGTAAAT -101
ATTTTTCTTTTTAATTCTAAATCAATCTTTTTCAATTTTTTGTTTGTATT -51
CTTTTCTTGCTTAAATCTATAACTACAAAAAACACATACATAAACTAAAA -1
PGK1-Promoter
CGATTTGGGCGCGAATCCTTTATTTTGGCTTCACCCTCATACTATTATCA -601
REB1 ABF1
GGGCCAGAAAAAGGAAGTGTTTCCCTCCTTCTTGAATTGATGTTACCCTC -551
ABF1
ATAAAGCACGTGGCCTCTTATCGAGAAAGAAATTACCGTCGCTCGTGATT -501
RAP1 GCR1
TGTTTGCAAAAAGAACAAAACTGAAAAAACCCAGACACGCTCGACTTCCT -451
GTCTTCCTATTGATTGCAGCTTCCAATTTCGTCACACAACAAGGTCCTAG -401
CGACGGCTCACAGGTTTTGTAACAAGCAATCGAAGGTTCTGGAATGGCGG -351
GAAAGGGTTTAGTACCACATGCTATGATGCCCACTGTGATCTCCAGAGCA -301
AAGTTCGTTCGATCGTACTGTTACTCTCTCTCTTTCAAACAGAATTGTCC -251
GAATCGTGTGACAACAACAGCCTGTTCTCACACACTCTTTTCTTCTAACC -201
AAGGGGGTGGTTTAGTTTAGTAGAACCTCGTGAAACTTACATTTACATAT -151
ATATAAACTTGCATAAATTGGTCAATGCAAGAAATACATATTTGGTCTTT -101
TCTAATTCGTAGTTTTTCAAGTTCTTAGATGCTTTCTTTTTCTCTTTTTT -51
ACAGATCATCAAGGAAGTAATTATCTACTTTTTACAACAAATATAAAACA -1
For Yeast genes there is also another database calles yeast promoter atlas, the sequences however differ from the first one (http://ypa.ee.ncku.edu.tw/) Here the sequences from YPA Media:Tum12Promotersequenzen.pdf, Media:Tum12 Promoterliste.pdf
Vectorsystems for Yeast
A common commercial Vector system for Yeast transformation is the pESC lineage from Stratagene
a broad range of Protocols for yeast transformation and for the making of competent cells can be found on [1] from several laboratories that work with yeast.
Plasmids from Publications as well as from companies can be viewed at[2] and can also be ordered there, as far as I could see they are far less expensive than those from companies.
 "
"