Team:TU Munich/Project/Caffeine
From 2012.igem.org
(→General remarks and issues) |
(→Biobricks and sequences) |
||
| Line 75: | Line 75: | ||
=== cDNA Synthesis === | === cDNA Synthesis === | ||
* If we do not want to order the sequences for the N- methyl transferases, another possibility would be an isolation of the corresponding genes by cDNA synthesis. Of course, it would be much more elaborate and i also do not know, wether it would be cheaper in the end. Anyway,[[Media:TUM12_Cloning_and_Characterization_of_N-Methyltransferases_Involvced_in_Caffeine_Biosynthesis.pdf| ''Uefuji et al. (2003)'']] show how they got their sequences. Interesting thing: Due to high sequence- homology of those three genes (partly up to 97%), one primer- pair is enough, to generate all three cDNAs, because their sequences (containing both, start and stop- codon) are absolutely identical in each gene. | * If we do not want to order the sequences for the N- methyl transferases, another possibility would be an isolation of the corresponding genes by cDNA synthesis. Of course, it would be much more elaborate and i also do not know, wether it would be cheaper in the end. Anyway,[[Media:TUM12_Cloning_and_Characterization_of_N-Methyltransferases_Involvced_in_Caffeine_Biosynthesis.pdf| ''Uefuji et al. (2003)'']] show how they got their sequences. Interesting thing: Due to high sequence- homology of those three genes (partly up to 97%), one primer- pair is enough, to generate all three cDNAs, because their sequences (containing both, start and stop- codon) are absolutely identical in each gene. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
=== General === | === General === | ||
Revision as of 14:10, 15 August 2012



Contents |
Background and principles
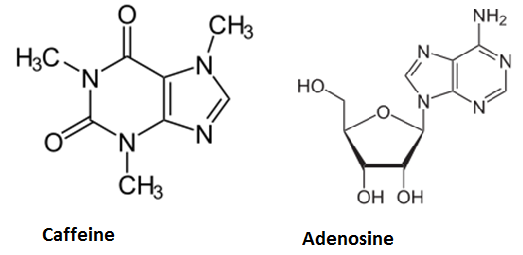
Caffeine is a purine- alkaloid and its biosynthesis is known in coffee plants and tea plants, for example. Its chemical structure is similar to the ribonucleoside adenosine. Hence it can block specific receptors in the hypothalamus in a competitive manner, which leads to decreased neurotransmitter- release and therefore decreased neuron activity. Biological background is to beware the brain of overexertion by inducing sleep and that is the reason for using coffeine to stay awake. On average, one cup of coffee contains about 50 - 130 mg Caffeine.
At higher doses (1g), caffeine leads to higher pulse rates and hyperactivity, but until that i think the beer would have done its work already...
Caffeine was shown to decrease the growth of E. Coli and Yeast reversibly as of a concentration of 0,1% by acting as a mutagen (Putrament et al., On the Specificity of Caffeine Effects, MGG, 1972), but previous caffeine synthesis experiments (see below) have only led to a concentration of about 5 µg/g (per g fresh weight of tobacco leaves), so i do not think we would reach the problematic concentration.
It has already been achieved to produce caffeine in tobacco plants ( Uefuji et al., 2005; Yun- Soo Kim et al., 2007 ), but has never been performed in yeast.
Biosynthesis
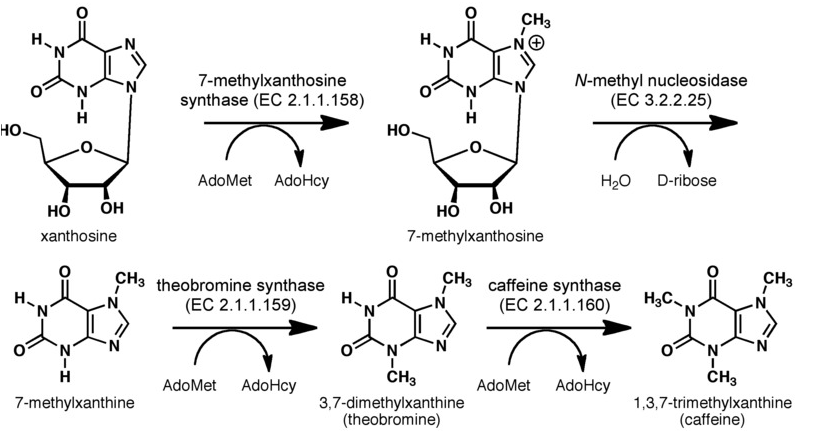
The biosynthetic pathway of caffeine (1,3,7 Trimethylxanthine) starts with xanthosine, which is a natural component of the purine- metabolism of all organism. Necessary for its production are three distinct N- methyl transferases and one nucleosidase, whereupon it has not been totally elucidated whether the nucleosidase reaction is catalyzed by any purine nucleosidase or by the first N- methyl transferase of the reaction cascade shown in the picture (but the latter assumption is favoured (H. Ashihara et al., 2008), because an in vitro synthesis of caffeine with the three N- methyl transferases has already been shown). I have indicated, that the caffeine syntase (last reaction step) can catalyze both, the conversion of 7- methyl xanthine to theobromine and the methylation of theobromine to caffeine. This is true, indeed, but Uefuji et al. (2003) showed, that the affinity to 7- methyl xanthosine is less than one sixth of that of CaMXMT1 (there are two isoformes of CaMXMT), so it is much better to express both enzymes. One can also see the Km values for the required enzymes in this paper - it shows that the substrate affinity decreases continiously towards the endpoint (caffeine), "making the reaction proceed irreversibly and stepwise" (Uefuji et al., 2003, p.377).
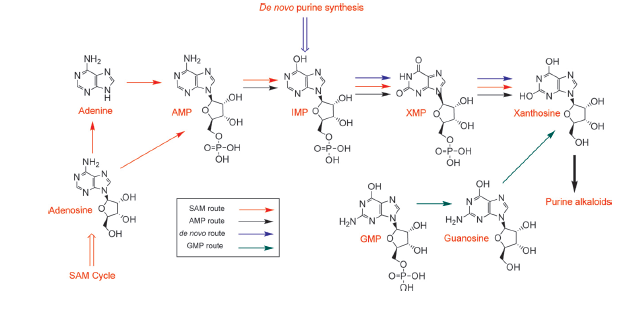
The chemical compound xanthosine is produced via at least four different routes, shown in the picture "xanthosine routes". To improve caffeine production, these pathways could be a possible target for metabolic engineering. Anyway it should be interesting/ necessary for this project to determine the in vivo xanthosine concentration of yeast.
Catabolism
Caffeine is demethylated to theophylline by 7N- demethylase (main pathway). The decreased rate of this reaction is the reason for the accumulating caffeine in the plant. Afterwards theophylline is degraded to xanthine via 3- methylxanthine and xanthine enters the conventional purine catabolism pathway (degradation to CO2 and NH3) (see H. Ashihara et al., 2008, p. 846). This catabolistic pathway is another possible target for metabolic engineering to increase the amount of caffeine (e.g. partially inhibition of 7N- demethylase (?))
Idea
The idea is to perform a heterologous gene expression of the distinct N-methyl transferases required for caffeine biosynthesis. The research groups which accomplished caffeine production in transgenic tobacco plants used the following three genes:
- CaXMT1 (AB048793)
- CaMXMT1 (AB048794)
- UniProt entry: Q9AVJ9
- 2.1.1.159
- This gene is eventually not essential, for CaDXMT1 is able to catalyze this reaction, too.
- Complete mRNA- sequence: [CaMXMT1] (coffea arabica)
- Coding sequence: bp 32 - 1168 ==> total length: 1137 bp
- Problematic restriction sites: EcoR1 at bp 722- 728
- CaDXMT1 (AB084125)
=== General ===:
In any case, we will have to order special RBS from the Partsregistry, for example [BBa_J63003], because they exhibit organism- specificity. However, an RBS is not essential in eukaryotic gene translation.
All mentionend methyltransferases use SAM als methyl- donor and are located in the cytoplasm of the plants. Furthermore they exist as homodimers, being also able to form heterodimers with each other (see BRENDA, also for further characteristics). The temperature and pH optimum of all three enzymes is quite similar between 20°C - 37°C and 7,5 - 8,5, respectively. (On a recent brewery guide tour i happend to learn that the pH of beer is slightly acid, but i do not know how much influence that would have on the enzyme activity).
General remarks and issues
Analytical Methods
- Detection and quantification of the produced caffeine can be performed by the use of HPLC. Uefuji et al., 2005; describe those and other relevant methods in this context.
- Proof of the gene- expression (of the necessary methyl- transferases) could be accomplished by standard methods (RT PCR and Western Blot, see also Uefuji et al., 2005; and Yun- Soo Kim and Hiroshi Sano, 2007;)
Gene Expression
- To increase gene expression it is possible to work with double promoter constructs. I am working with that in my bachelors thesis and my adviser said this would be an often underrated possibility to improve gene expression. Perhaps we can make use of this.
cDNA Synthesis
- If we do not want to order the sequences for the N- methyl transferases, another possibility would be an isolation of the corresponding genes by cDNA synthesis. Of course, it would be much more elaborate and i also do not know, wether it would be cheaper in the end. Anyway, Uefuji et al. (2003) show how they got their sequences. Interesting thing: Due to high sequence- homology of those three genes (partly up to 97%), one primer- pair is enough, to generate all three cDNAs, because their sequences (containing both, start and stop- codon) are absolutely identical in each gene.
General
Theoretically, we only need the coding sequences of the genes, because we ought to use the special RBS and Terminator biobricks from the registry and thus do not need the existing ones (in the 5' UTR and 3' UTR, respectively) (Anyway, they would probably not work in yeast)
Finally ordered Sequences
The ordered sequences were constructed as follows:
- the 5' UTR and 3' UTR of the sequences above were removed
- the yeast consensus sequence for improved ribosome binding (TACACA) was added 5' of the start codon ATG
- according to n- end rule and the yeast consensus sequence for improved ribosome binding, the first triplet after ATG (GAG) was exchanged with TCT (serine), to optimize both, protein stability and mRNA translation. This decision was made after proofing the 3D- structure of the enzyme CaDXMT1. Due to the fact, that the the first two residues of the amino acid sequence are not shown in the crystalized structure (probably because of high flexibility), Prof. Skerra said we can risk the exchange of this amino acid, for it is probably not that necessary for the uptake of the ligands (uniprot entry further shows, that it is not immediately involved in ligand binding in one of the three enzymes). Because of the high similarity of the enzyme- sequences, we also exchanged this amino acid in the enzyme CaMXMT1, although here is no 3D- structure available
- we added a c- terminal strep-tag for purification
- the remaining coding sequence was extended with the standard RFC10 prefix and suffix, respectively
- at last we made an optimization of the coding sequences with respect to the codon usage and mRNA structures (online tool of a gene- synthesis company)
- annotated sequences:
References
- Putrament et al., On the Specificity of Caffeine Effects, MGG, 1972
- H. Uefuji et al., Plant Physiology, 2003, Vol. 132, pp. 372–380
- H. Uefuji et al., Plant Molecular Biology, 2005, Vol. 59, p. 221–227
- H. Ashihara et al., Phytochemistry, 2008, Vol. 69, p. 841–856
- Yun-Soo Kim, Hiroshi Sano, Phytochemistry, 2008, Vol. 69, p. 882–888
 "
"