Team:TU Darmstadt/Protocols/GC-MS
From 2012.igem.org
(→GC-MS Purge and Trap) |
(→Development of a brand new method) |
||
| Line 55: | Line 55: | ||
[[File:SIM_Scan_optimal_method.jpg |900px| GC-MS spectra |center ]] | [[File:SIM_Scan_optimal_method.jpg |900px| GC-MS spectra |center ]] | ||
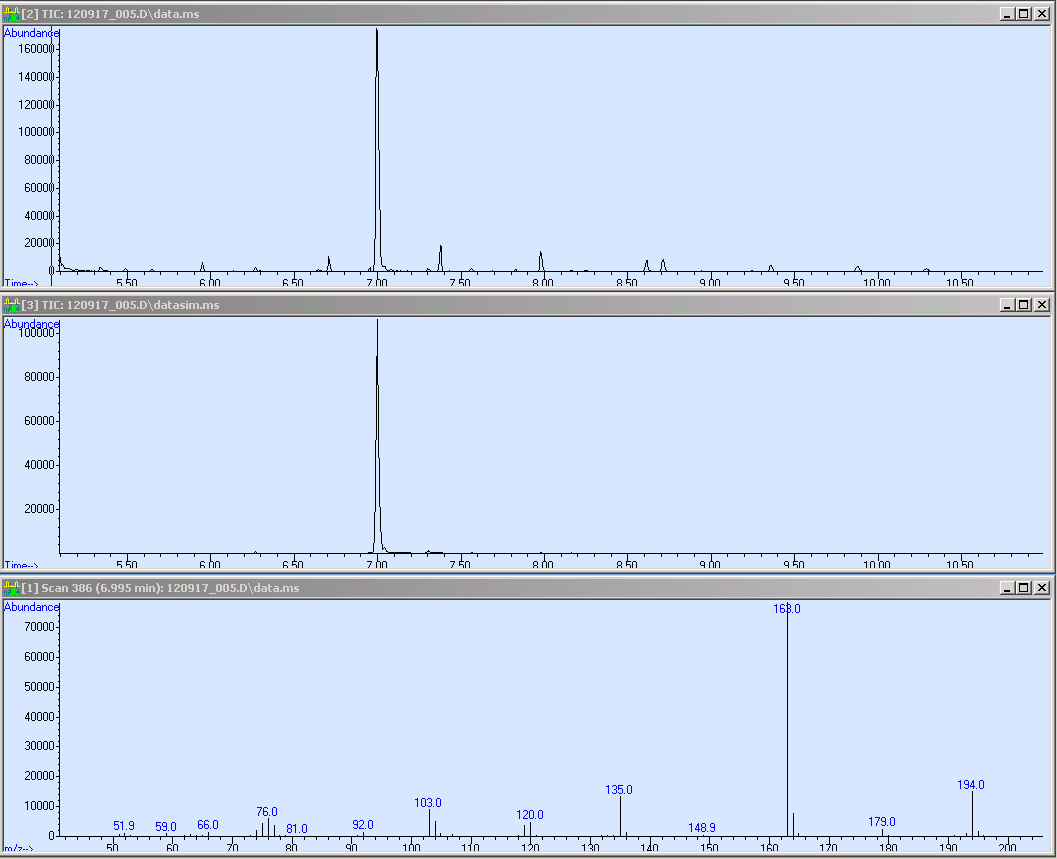
| - | The first window shows the | + | The first window shows the SCAN of the TPA reference sample. Notably the big peak at a retention time of about 7 minutes. The second window shows the single ion monitoring (SIM). For the SIM we used the mass of 163 as marker. The third is the mass spectra of the peak by 7 minutes. Hence it shows the characteristic fragmentation pattern of TPA-dimethylester. |
We came down on the following approach (see protocol below) where we extract and purified our substances out of the supernatant. Therefore, we have to transform TPA into the corresponding di-methyl-ester : | We came down on the following approach (see protocol below) where we extract and purified our substances out of the supernatant. Therefore, we have to transform TPA into the corresponding di-methyl-ester : | ||
| Line 61: | Line 61: | ||
Here we illustrate the reaction-mechanism of the ester reaction that we observed and thus used for our sample preparation. | Here we illustrate the reaction-mechanism of the ester reaction that we observed and thus used for our sample preparation. | ||
| - | So we developed the new sample | + | So we developed the new sample preparation protocol, we hope it will work. |
[Results] | [Results] | ||
Revision as of 00:28, 26 September 2012
Contents |
Theory
Gas chromatography mass spectrometry (GC-MS)
To validate that our designed bacteria is able to digest PET and convert it into chemicals which can be used by the organism, we decided to use the "gold standard" for substance identification gas chromatography-mass spectrometry. The gas chromatograph (GC) separates all substances via a purification column, afterwards the exact mass of each purified substance is measured by mass spectrometer. This gives a clearly proof of the exact amount of the substance which are produced by our designed organism. The difficulty in achieving good measurements of terephthalic acid (TPA), the main component of PET, is given by its physical properties. The aromatic body carries hydrophobic properties, while the two acid groups in para position are hydrophylic, this ambivalent character makes it a challenge to purify TPA by conventional purification columns. Since we searched various hours for a GC-MS protocol, we came to the conclusion that nobody did this before. That's why we first had to develop a new experimental setup to prepare our probes for the analysis.
GC-MS Purge and Trap
Thus we dont know in which matrix we can detect our analyte we try simultaneously a second approach. The purge and trap method is used to get an anaylte out of a water sample. Therfore, we purge our sample with an inert gas. Moreover, we trap our analyte with an organic trap as well as water with a water-trap. While gassing out the whole sample with the inert gas, the anaylte accumumlate on the trap. After that, we release the analyte from the trap with an temperature shift. Now, our analyte is ready to pass the column for the analysis.
Development of a brand new method
To measure TPA we have to embed it into a an appropriate matrix. One requirement to a matrix is that it has to be free from water. Hence we tested aceton. methanol and cycl. hexan. Unfortunately, TPA was only soluble in methanol (0,1 g/l). In our case, a matrix like cycl. hexan would be easier for the extraction of TPA out of our water samples. For our measurement approach we choose the following GC-Parameter
The first window shows the SCAN of the TPA reference sample. Notably the big peak at a retention time of about 7 minutes. The second window shows the single ion monitoring (SIM). For the SIM we used the mass of 163 as marker. The third is the mass spectra of the peak by 7 minutes. Hence it shows the characteristic fragmentation pattern of TPA-dimethylester. We came down on the following approach (see protocol below) where we extract and purified our substances out of the supernatant. Therefore, we have to transform TPA into the corresponding di-methyl-ester :
Here we illustrate the reaction-mechanism of the ester reaction that we observed and thus used for our sample preparation. So we developed the new sample preparation protocol, we hope it will work. [Results]
Protocol GC-MS sample preperation
- set up E.Coli culture (insert your experimental setup, e.g.with Transporter operon) 1.5 ml
- inducing (arabinose)
- growth
- spin down
- isolate the supernatant and
- dry it overnight on a heating-block
- mix with methanol 5ml
- add. 37% HCL 500µl
- incubate over night
- precipitation of proteins with 3% TCA (Trichlor Acectic Acid)
- wash with cyclohexane 4ml
- use only 3 ml (prevent water contamination) of the cyclohexane fraction
- ready for GC-MS analysis
Protocol GC-MS Purge and Trap sample preperation
- set up E.Coli culture (insert your experimental setup, e.g.with Transporter operon)
- inducing (arabinose)
- growth
- spin down
- isolate the supernatant and mix with methanol
- add. a fraction (eg. 10-50 µl ) to 10 ml water in a vial
Here you can see our [results]
 "
"