Team:TU Darmstadt/Modeling Docking
From 2012.igem.org
(→Goal) |
|||
| Line 53: | Line 53: | ||
===Theory=== | ===Theory=== | ||
===Goal=== | ===Goal=== | ||
| - | While PET.er is degrading PET into its monomers various chemical compounds like polymer plasticizer are released. | + | While PET.er is degrading PET into its monomers various chemical compounds like polymer plasticizer are released. Since PET is itself a waste material, it will never be in a pure state. Due to this, it is important to study possible interaction of plasticizer as well as PET with the degradation machinery. Hence an experimental design will give a deeper understanding of the interaction of our enzymes with additives. |
===Methods=== | ===Methods=== | ||
| - | Due to the absence of forcefield terms of plasticizer we choose a global docking approach to study the interaction. For the docking simulations we uses the Autodock plugin of Yasara structure. Autodock is one of the mreeost cized and scientific programs for Docking simulations. | + | Due to the absence of forcefield terms of plasticizer we choose a global docking approach to study the interaction. For the docking simulations we uses the Autodock plugin of Yasara structure. Autodock is one of the |
| + | ?? mreeost cized and scientific programs for Docking simulations. ?? | ||
===Docking Protocol=== | ===Docking Protocol=== | ||
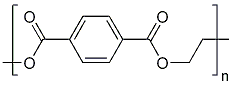
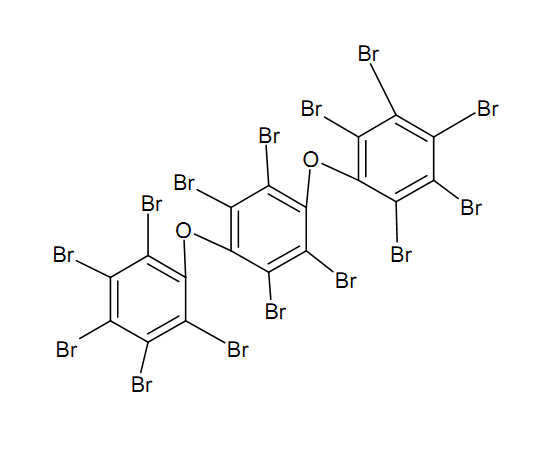
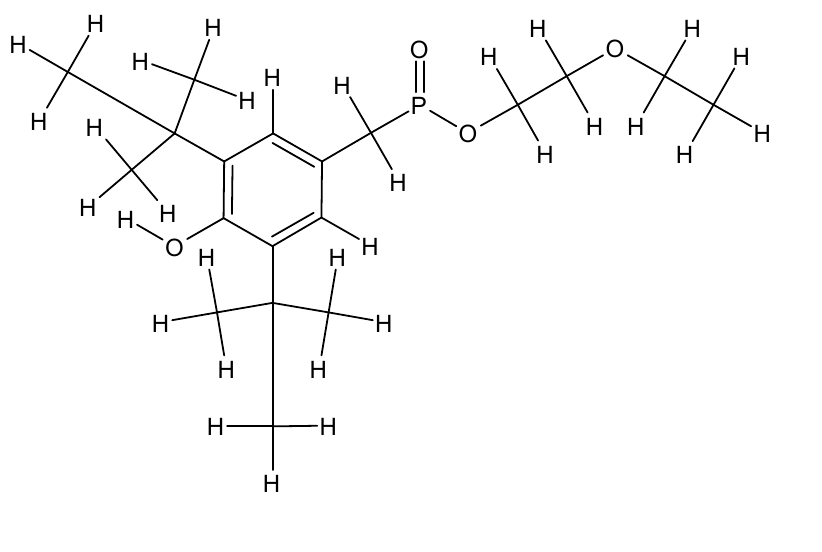
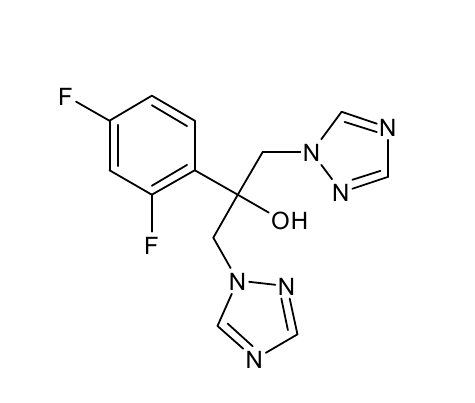
| - | For the docking approach we utilized the standard protocol dock_run.mcr and parallelized it up to six processors. Moreover, we | + | For the docking approach we utilized the standard protocol dock_run.mcr and parallelized it up to six processors. Moreover, we choAse three additives out of the literature acting as the ligand within the simulations. |
*OET-dimeric | *OET-dimeric | ||
[[File: PET.png |250px|center]] | [[File: PET.png |250px|center]] | ||
| Line 73: | Line 74: | ||
*Flexible Ligand | *Flexible Ligand | ||
===Analytics=== | ===Analytics=== | ||
| - | We extract binding energy from the atomic | + | We extract binding energy from the atomic B-factors and the binding constant from the Atomic property derived from all our calculations. We used scatter plots with the binding constant and the inhibition constant in order to compare the plasticizer as well as the PET substrat |
===Results=== | ===Results=== | ||
Revision as of 08:52, 23 September 2012
| Homology Modeling | | Gaussian Networks | | Molecular Dynamics | | Information Theory | | Docking Simulation |
|---|
Contents |
Docking
Theory
Goal
While PET.er is degrading PET into its monomers various chemical compounds like polymer plasticizer are released. Since PET is itself a waste material, it will never be in a pure state. Due to this, it is important to study possible interaction of plasticizer as well as PET with the degradation machinery. Hence an experimental design will give a deeper understanding of the interaction of our enzymes with additives.
Methods
Due to the absence of forcefield terms of plasticizer we choose a global docking approach to study the interaction. For the docking simulations we uses the Autodock plugin of Yasara structure. Autodock is one of the ?? mreeost cized and scientific programs for Docking simulations. ??
Docking Protocol
For the docking approach we utilized the standard protocol dock_run.mcr and parallelized it up to six processors. Moreover, we choAse three additives out of the literature acting as the ligand within the simulations.
- OET-dimeric
- S-120
- AO-8
- TPF
Parameter for the Docking Experiment
- Ligands = S120, AO-8, TPF, dimer OET
- Number of docking runs = 200
- ForceField AMBER03
- Flexible Ligand
Analytics
We extract binding energy from the atomic B-factors and the binding constant from the Atomic property derived from all our calculations. We used scatter plots with the binding constant and the inhibition constant in order to compare the plasticizer as well as the PET substrat
 "
"