Team:Stanford-Brown/HellCell/Plasmid
The Test Plasmid
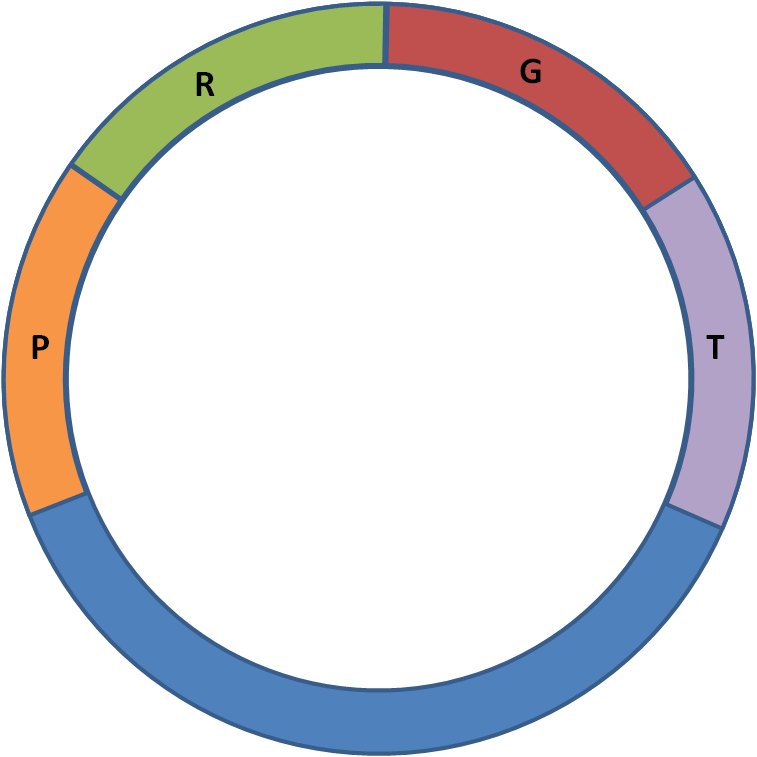
To test the genes that we isolated from various organisms in hopes of arming our Escherichia coli against the extremes, we used standard BioBrick assembly to place all of the genes into the same "Test Plasmid." We used a strong promoter and ribosome binding site (RBS) for high protein expression, increasing the chances of visible and statistically significant results. Instead of a regular terminator, we used GFP-producing part BBa_E0840 (that contains a RBS, GFP, and a double terminator) so that we could have an easy visual confirmation that transcription and translation of that region is occurring (See Figure 1). Because of the way that we assembled the parts together, all of these composite parts ended up on psB1A2 (the plasmid BBa_E0840 was on).
Figure 1: Test Plasmid, inserting one gene
P = Strong Promoter, BBa_J23100 BBa_J23100
R = Strong RBS, BBa_B0030
G = Gene of interest
T = GFP Reporter, BBa_E0840
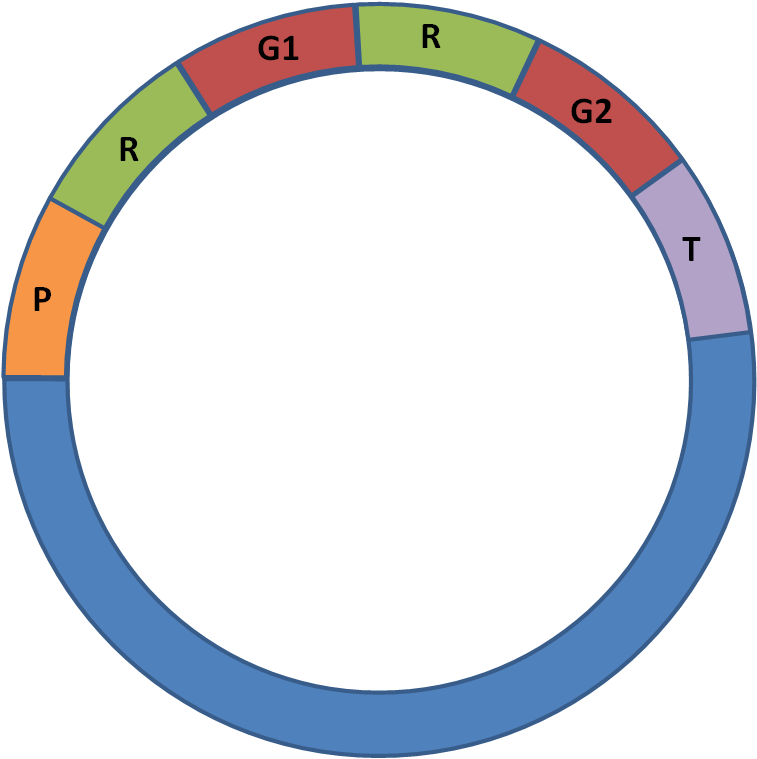
In some cases, we wished to insert two genes into the same plasmid as they both contributed to one pathway. We used the following scheme:
Figure 2: Test Plasmid, inserting two-gene pathway
P = Strong Promoter, BBa_J23100
R = Strong RBS, BBa_B0030
G1 = Gene of interest, first step of pathway
G2 = Gene of interest, second step of pathway
T = GFP Reporter, BBa_E0840
Our negative control for all of our assays were NEB 5α cells transformed with an empty psb1A2 plasmid.
 "
"