Biosynthesis - Violacein
Violacein is a purple chromobacterial pigment produced from tryptophan. In its biosynthetic pathway there are 3 possible branches and 2 side products. The five key enzymes of Chromobacterium violaceum could be expressed in E.Coli and corresponding parts are available in the Registry. The Cambridge 2009 iGEM team had succesfully achieved the production of violacein.
Our membrane rudder calls for a branched reaction to test its efficiency. We wanted to examine the advantages of membrane system in adjusting quantity of different products.
We modified the five vio parts and inserted them in the membrane assembly, producing a plasmid with violacein biosynthetic enzyme, light-inducing protein as well as a membrane protein. Analysis of bacteria extracts showed a satisfying decrease in side products between light-induced samples and the control group. These results demonstrate a rather promising way of using membrane rudder to switch the direction of reaction.
Background
Violacein is a pigment produced by several bacteria through pathway of 5 related genes, vioA, vioB, vioC, vioD and vioE. It was initially discovered in Chromobacterium violaceum. This metabolite of tryptophan has such special applications as antibacterial, anti-trypanocidal, anti-ulcerogenic, and anticancer drugs.
There are two interesting branches in this pathway which could be utilized in service of verification of the membrane rudder. We can identify the different end-products by means of separation, such as thin-layer chromatography (TLC), or high-performance liquid chromatography (HPLC). The product proportion change demonstrates the efficiency of alteration in reaction direction.
Violacein Biosynthetic Pathway
The violacein biosynthetic pathway includes 5 key enzymes which work in conjunction. VioA, a flavoenzyme and VioB, a heme protein, they work together to oxidize and dimerize tryptophen into IPA imine dimer. Then vioE induces a indole rearrangement,producing prodeoxyviolacein, also known as PVA.
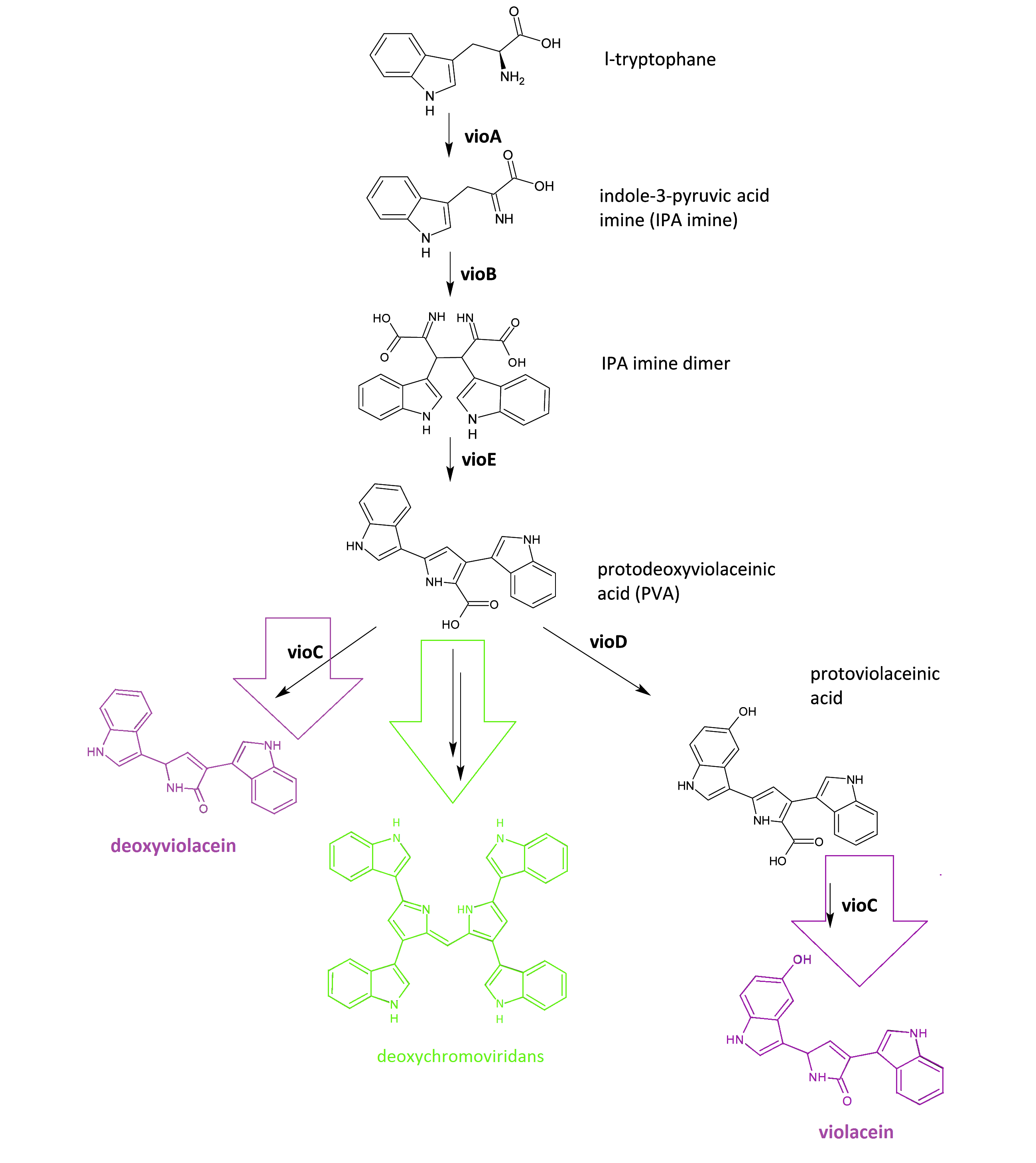 Fig.1 :The violacein biosynthetic pathway. The purple part on the left indicates the branch of pigment deoxyviolacein.The violet part on the right indicates the branch of pigment violacein. The green part indicates the branch of pigment deoxychromoviridans.
One key prerequisite in the metabolic flux is that: Automatically VioA, VioB and VioE can assemble and function to produce PVA. There exists one intrinsic E.coli enzyme that aids in an additional side reaction, further modifying PVA into a green pigment called deoxychromoviridans(1st main product).
The last two proteins, VioC and VioD are flavin-dependent oxygenases. VioC alone transfers PVA into a purple pigment named deoxyviolacein(2nd main product), while vio C could also act sequentially with vioD.
vioD hydroxylates 5-position indole ring, then the other 2-position indole ring is processed by VioC to create the oxindole, and in this way violacein(3rd main product) is produced.
Design of Membrane Assembly
Theoretically our membrane rudder offers a method to fine-tune reaction direction, and in the meantime decrease side-products quantity.
Control over reaction flow
Since VioA,VioB and VioE normally aggregate to finish the transfer from tryptophen to PVA, the key to direction of the rest part of pathway is if PVA could be passed on to vioD timely. We intergrated M1-vioA with vioC, and M3-vioE with vioD to assure the PVA would be catalyzed by the next enzyme right away.Through connecting VIVID split protein and VioC and VioD seperately, a light signal could efficiently shorten distance between these two enzymes. As long as VVD is photo-induced and dimerized, they could act like magnets that pull vioC and vioD together, facilitating the indole ring rearrangement under catalization of VioC and VioD,in this case violacein should be the dominant product we expect. Since vioB and vioE normally function in dimer state, pRSFduet with rbs-vioB and rbs-vioE was transformed into E.Coli to ensure dimerization, then all five functional enzymes could be expressed now.
To quantify the practical effect of photo-inducing direction alteration, we set a control group and conducted experiments as designed.There was no other difference but one that the control group was induced under absolutely no light signal.
figure2//膜蛋白图片
Suppression over side-reactions
According to the mechanism behind our design of membrane rudder, a considerable reduction of side-product is expected. Under light signal, VioD and VioC get close to other Vio proteins,the assembly of key enzymes near the membrane also lessens the proportion of dissociative intermediates and makes them easier to be captured by VioD and VioC. The overall amount of available tryptophen in a bacterium is fixed, preferance to the production of violacein would inevitably cut the supply to the other branches of reaction.
In a word, productivity of side-products will be decreased and tendancy of low production is possible.
To verify our assumption, rbs-vio genes were also inserted in the three modified plasmids and transformed into E.Coli.
Results and Discussion
Overview
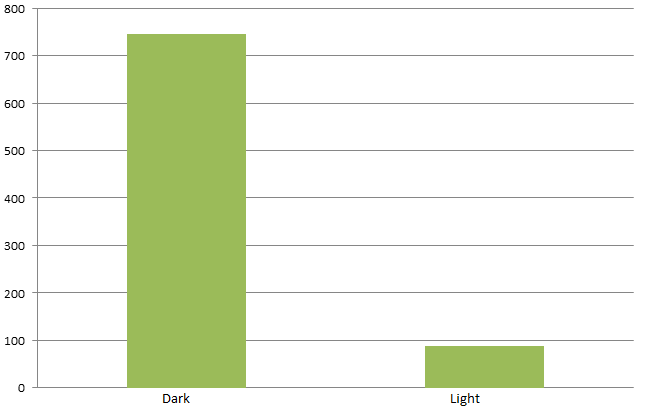
By attaching the core enzymes to the VIVID, we succeeded to control the flow of branched chain reaction, such as violacein biosynthetic pathway. Results indicate that deoxybiolacein was almost absent from the extracts of bacterial cultures induced by light while samples restrained from light containd deoxyviolacein.(Figure X, compare X and X) Moreover, a significant decrease of side products such as deoxychomoviridans was detected, when membrane complex system was present. (Figure X, compare lane X and X)
Switch Analysis
The switch of the reaction can be realized by the extracellular signals. Through the light induction, the reaction producing deoxyviolacein by vioC alone is inhibited due to the lack of interaction with PVA. On the other hand, as long as light is restrained from the bacteria, however, the above reaction is initiated leading to the production of deoxyviolacein.
From the HPLC, we find out that under the induction of light, production of deoxyviolacein was inhibited (less than 1%) while most products were violacein. Without the light induction, however, the pathway leading to the deoxyviolacein was initiated and some deoxyviolacein was produced(about 8%). Such results confirmed our membrane rudder system and indicated that by controlling the extracellular signal, such as light, we are able to manipulate the branched chain reactions thus producing the target products we desire.
Inhibition of side reaction
membrane complex system would help to reduce the amount of side-products to some extent. Under the case that the enzymes involved in the side reaction are situated in the cytoplasm, they are less competent than the core enzymes attached on the membrane to mediate the subsequent reactions due to the spatial obstacle.
From the HPLC results, we detected a distinct decrease of side-products deoxychomoviridans. Such experiment results coincided with our anticipation and demonstrated the significance membrane complex system has in the reduction of side-products.
Reference
1. Balibar, C. J. and C. T. Walsh (2006). "In vitro biosynthesis of violacein from L-tryptophan by the enzymes VioA-E from Chromobacterium violaceum." Biochemistry 45(51): 15444-57.
2. Hoshino, T. "Violacein and related tryptophan metabolites produced by Chromobacterium violaceum: biosynthetic mechanism and pathway for construction of violacein core." Appl Microbiol Biotechnol 91(6): 1463-75.
3. Shrode, L. B., Z. A. Lewis, et al. (2001). "vvd is required for light adaptation of conidiation-specific genes of Neurospora crassa, but not circadian conidiation." Fungal Genet Biol 32(3): 169-81.
|  "
"