Membrane Rudder - Violacein & Deoxyviolacein synthetic pathway
Dynamically and artificially regulating the direction of biochemical pathway in vivo has remained a challenge for scientists. We are now to achieve this goal through controlling the aggregation state of different enzymes in branched biological reactions based on Membrane Scaffold. We named this universal device Membrane Rudder.
Violacein is a purple chromobacterial pigment synthesized from the basic amino acid tryptophan. Its biosynthetic pathway contains 3 branches and produces 3 final products, which are violacein, deoxyviolacein and deoxychromoviridans respectively. Thus it is incorporated to test the feasibility of Membrane Rudder
To switch the direction of Violacein and deoxyviolacein synthetic pathway through light signal
To switch the direction of Violacein and deoxyviolacein synthetic pathway through RNA signal
To justify the universality of Membrane Rudder
√ Successfully switched the direction of Violacein synthetic pathway by light signal
√ Decreased yields of side product deoxychromaviridans by 8-fold with Membrane Rudder compared to group with free cytoplasmic enzyme
√ Constructed a device that connects genetic circuits to post-translational Membrane Scaffold system by recruiting RNA as a controlling signal.
Background
Violacein is a violet pigment produced by several bacteria through pathway of 5 related genes, vioA, vioB, vioC, vioD and vioE. It was initially discovered in Chromobacterium violaceum. This metabolite of tryptophan has such special applications as antibacterial, anti-trypanocidal, anti-ulcerogenic, and anticancer drugs.
There are three branches in this pathway which could verify the efficiency of the Membrane Rudder. We could identify the different end-products by high-performance liquid chromatography (HPLC). The change of products proportion will demonstrate the efficiency of direction alteration .
Violacein Biosynthetic Pathway
The violacein biosynthetic pathway includes 5 key enzymes which work in conjunction. VioA, a flavoenzyme and VioB, a heme protein, work together to oxidize and dimerize tryptophen into IPA imine dimer. Then VioE induces an indole rearrangement, producing prodeoxyviolacein acid, also known as PVA.
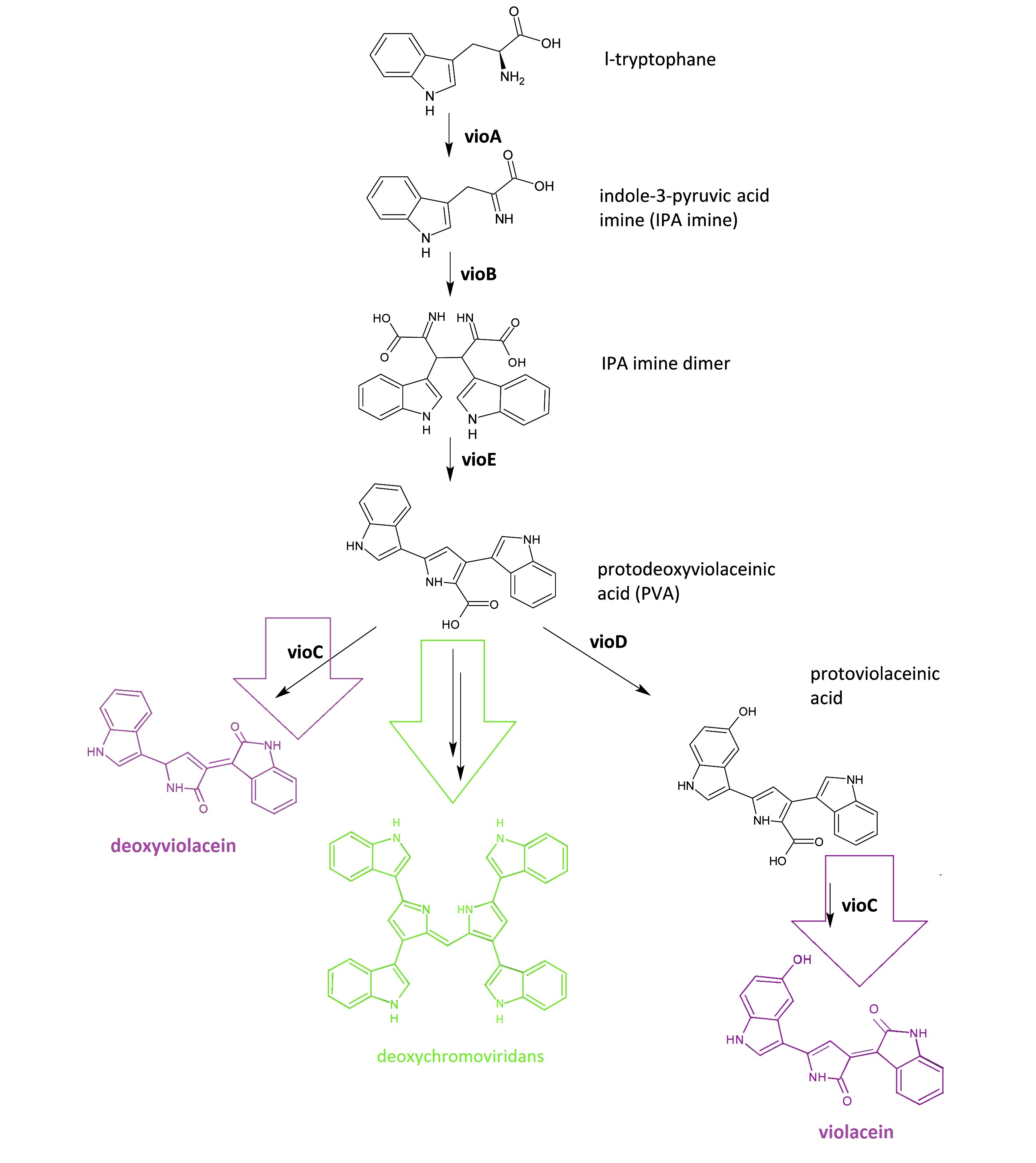 Fig.1 :The violacein biosynthetic pathway. The purple part on the left indicates the branch of pigment deoxyviolacein.The violet part on the right indicates the branch of pigment violacein. The green part indicates the branch of pigment deoxychromoviridans.
The main intermediate, PVA now is produced by the sequential catalyzation of VioA, VioB, VioE, then branches are about to appear.
There exists one intrinsic E.coli enzyme that automatically contribute to an additional side reaction, further modifying PVA into one green pigment called deoxychromoviridans.
The last two enzymes, VioC and VioD are flavin-dependent oxygenases. VioC alone transforms PVA into a purple pigment called deoxyviolacein, the second product, while it could also act cooperatively with VioD. The 3rd branch works like this: VioD hydroxylates 5-position indole ring, then the other 2-position indole ring is processed by VioC to create the oxindole. It is in this way could violacein be produced.
Design of Membrane Assembly
Theoretically our Membrane Rudder offers a method to fine-tune reaction direction.
Switch of reaction direction
We constructed our device as demonstrated in Fig.2. In Violacein Membrane Rudder System, VioA, VioB, VioE and VioC constitutively aggregate. When blue light is present, VioD with Membrane Anchor would aggregate with complex of VioA,B,C and E, making violacein the dominant final product.But if incubation is carried out in darkness, VioD with Membrane Anchor won't associate with VioA,B,C and E. Thus the biosynthetic pathway for deoxyviolacein is switched on.
For VioB and VioE only function normally in dimereic state, free VioB and free VioE were coexpressed with membrane assembly carrying VioA,B,C,D and E to ensure the normal function of the whole system.
To characterize the practical effect of Violacein Membrane Rudder System, we coexpressed all proteins in Fig.2 with free VioB and VioE. Bacteria in experimental group were induced at a L-Arabinose concentration of 0.1% under blue light while control group were induced in the dark with other conditions all the same.
The BioBrick Part VVD-MA5-vioC and VVD-MA6-vioD is [http://partsregistry.org/Part:BBa_K771205 Part:BBa_K771205] and [http://partsregistry.org/Part:BBa_K771206 Part:BBa_K771206], respectively. For Membrane Anchor 2-vioA, MA3-vioB, MA4-vioE, the corresponding part is [http://partsregistry.org/Part:BBa_K771201 Part:BBa_K771201], [http://partsregistry.org/Part:BBa_K771202 Part:BBa_K771202], [http://partsregistry.org/Part:BBa_K771203 Part:BBa_K771203], respectively.
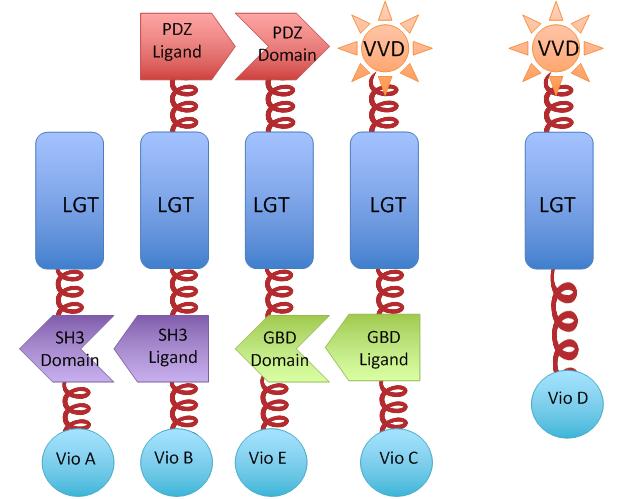 Fig.2 :The violacein Rudder construction. Suppression of side-product
According to the mechanism behind our design of membrane rudder, a considerable reduction of deoxychromoviridans production is expected. Under light signal, VioD and VioC get close to other Vio enzymes. The assembly of sequential enzymes helps substrate flow to downstream enzymes, making violacein more preferably synthesized . The overall amount of available tryptophan in a bacterium is fixed, so preference to the production of violacein would inevitably cut the supply to the other branches of reaction.
In a word, productivity of deoxychromoviridans will be decreased.
To verify our assumption, free VioABCDE genes are also transformed into E.coli as control group.
Results and Discussion
Overview
By attaching the core enzymes to the VIVID, we managed to control the flow of branched chain reaction, such as violacein biosynthetic pathway. Results indicate that the extracts of bacterial cultures induced by light contained almost no deoxyviolacein while it was produced in the samples restrained from light. Moreover, a significant decrease of side products such as deoxychomoviridans was detected when membrane complex system was present.
Switch Analysis
The switch of the reaction can be realized by the extracellular signals. Through the light induction, the reaction producing deoxyviolacein by VioC alone is inhibited due to the lack of interaction with PVA. On the other hand, as long as light is restrained from the bacteria, however, the above reaction is initiated leading to the production of deoxyviolacein.
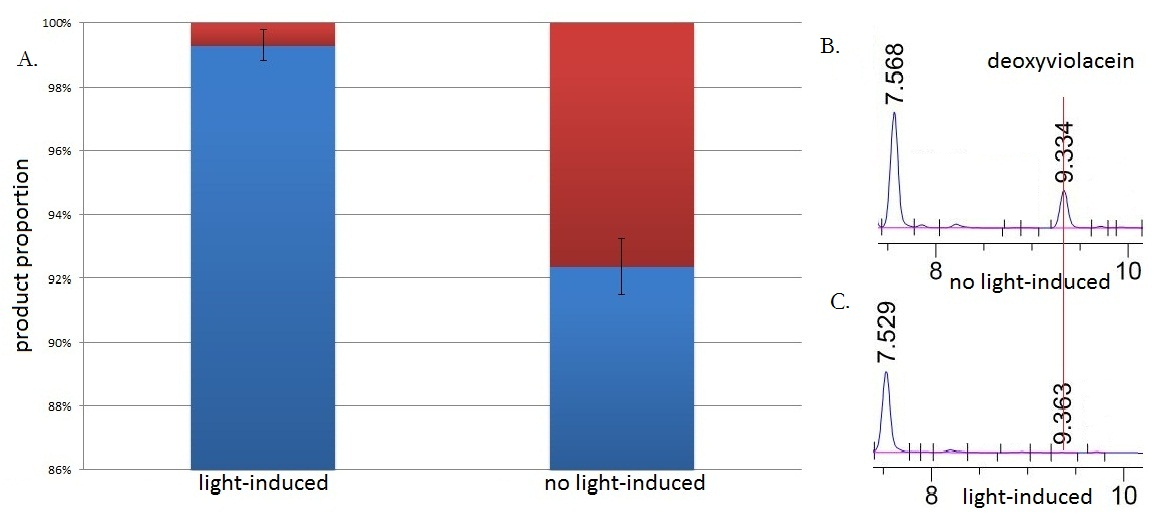 Fig.3 :HPLC result (right) and the ratio(left) of deoxyviolacein in sample with light induction and without light induction. Fig. B represents the deoxyviolacein HPLC peak results from sample with no light induction. Fig. C represents the deoxyviolacein HPLC peak results from sample with light induction. Results indicate that the amount of the deoxyviolacein from the sample without light induction is tremendously larger than that from the sample with light induction. The red part in the Fig.A represents the portion of deoxyviolacein and the blue part represents violacein We ran an HPLC (SHIMADZU LC-20AP,C18 reversed column) test with the purple samples extracted from the bacterium culture.Then we ran an mass spectrometry (Thermo Ultra GC-ISQ) test and verified the corresponding molecule role of every main peak.
The HPLC results show that the peak of deoxyviolacein appears at about 9 minutes after the injection. We find out that under the induction of light, production of deoxyviolacein was inhibited (to almost 0%) while most products were violacein. Without the light induction, however, the pathway leading to the deoxyviolacein was initiated and thus deoxyviolacein was produced(about 8%). Such results confirmed our membrane rudder system and indicated that by controlling the extracellular signal, such as light, we are able to manipulate the branched chain reactions, thus producing the target products we desire.
Suppression of side reaction
Membrane complex system would help to reduce the amount of side-products to some extent. Under the case that the enzymes involved in the side reaction are situated in the cytoplasm, they are less competent than the core enzymes attached on the membrane to mediate the subsequent reactions due to the spatial obstacle.
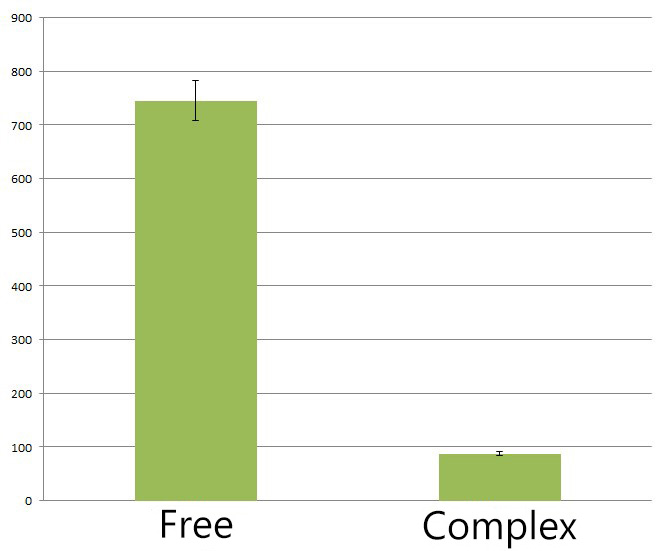 Fig.4 :The HPLC peak area of deoxychromoviridans from sample with light induction and free vioABCDE control group. From the HPLC results, we detected a distinct decrease of side-product, deoxychomoviridans. Judging from the peak area, we find out that the amount of deoxychromoviridans from rbs-vio control group is nearly eight times that from light-induced group. Such experiment results coincided with our anticipation and demonstrated that the membrane complex system would enhance the usage of substrates and thus reduce the amount of intermediate products and other subsequent side-products.
Now Trying
Multiple choices for signal control over reaction is the next target for membrane rudder, so we constructed a modified set of membrane assemblies where a molecule named RNA D0 offered another way to regulate direction.
As illustrated in the Construction section, we fused MS2 and PP7 aptamer domain to membrane assembly 1 and membrane assembly 2 correspondingly, and they are the cognate binding domains of a RNA molecule we found named RNA D0. If the RNA D0 gene is set behind a genetic circuit, its transcription can be regulated by the circuit and on the other hand, will lead to aggregation of membrane assembly 1 and 2.
If VVD and RNA D0 are put in the system together,they could aid in realizing free switch between 2 or 3 branches of reaction.
More overwhemingly, theRNA D0 modified system establish a connection between genetic circuit (in other words,traditional control mode on transcription level) and membrane rudder(our novel dynamic control system on protein level).
We have constructed the PP7 and MS2 standardized BioBrick part, [http://partsregistry.org/Part:BBa_K771111 Part:BBa_K771111] and [http://partsregistry.org/Part:BBa_K771112 Part:BBa_K771112], respectively.
All necessary parts have been constructed and submitted to the registry while relating confirmatory experiments are being conducted.
Reference
1. Balibar, C. J. and C. T. Walsh (2006). "In vitro biosynthesis of violacein from L-tryptophan by the enzymes VioA-E from Chromobacterium violaceum." Biochemistry 45(51): 15444-57.
2. Hoshino, T. "Violacein and related tryptophan metabolites produced by Chromobacterium violaceum: biosynthetic mechanism and pathway for construction of violacein core." Appl Microbiol Biotechnol 91(6): 1463-75.
3. Shrode, L. B., Z. A. Lewis, et al. (2001). "vvd is required for light adaptation of conidiation-specific genes of Neurospora crassa, but not circadian conidiation." Fungal Genet Biol 32(3): 169-81.
|  "
"



