Membrane Rudder
Background
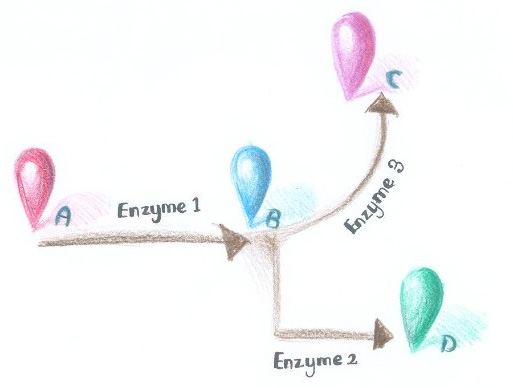 Fig.1 :Demonstration of branched reactions. When signal that induces the dimerization of Enzyme Complex 1 and 2 is present, they would dimerize, making product D more dominant. On the contrary, when signal that induces the dimerization of Enzyme Complex 1 and 3 is present, they would aggregate, so product C is more dominant. Membrane Accelerator employed constitutively dimerizing proteins to assemble enzymes. Imagine if we replace those constitutively dimerizing proteins with signal-induced dimers, which could only form dimers with signal input, then it is possible to dynamically control the direction of branched reactions.
Finding a signal inducible dimer is not difficult. There are indeed many proteins that could behave like that.
Light Signal
 Fig.8 :Dark state VVD dimers, from PDB ID 2PD7 Light is an intriguing signal to regulate E.coli activity because it is easy to obtain, highly tunable, nontoxic. A light-switchable system could be very fascinating. So we decide to use light as a inducing signal that could control the assembling of our Membrane Rudder system.
We recruited Vivid(VVD) protein, photoreceptor of Neurospora crassa and can form dimer in the presence of blue light and disassociate as light is off. The VVD belongs to the Per-Arnt-Sim(PAS) protein superfamily.
 Fig.9 :Fusion protein constructed to test the usability of VVD protein in our membrane system
VVD mutant (C71V and N56K) is harder to dimerize in the dark and easier to disassociate under blue light compared with wildtype VVD. Still, we used split GFP fluorescence complementation assay to test the system.
Transcription Signal
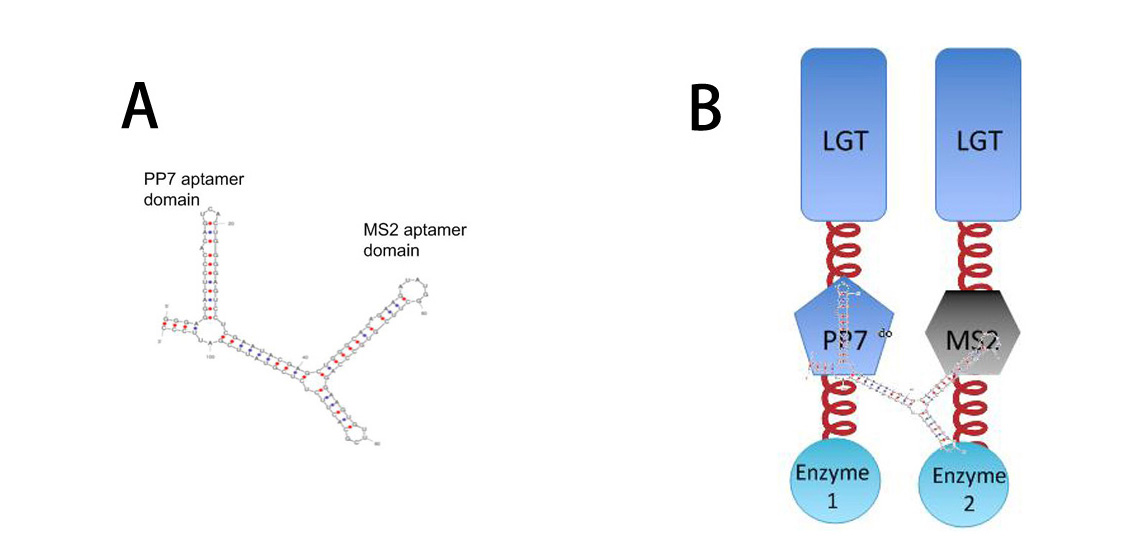 Fig.10 :A: Sketch of signal RNA molecule which consists of PP7 and MS2 aptamer domains. This RNA molecule is called PM in our system B: Through fusing PP7 and MS2 protein to our membrane device, PM (RNA molecule in Figure A) can function as a bridge to connect different membrane scaffold. So far, our system is a generally about posttranslational control over the host cell. To connect our relatively isolated system to transcription level, we employed RNA signal, which is present in cytoplasm and let our system under control at transcription level. When certain RNA molecule with dimerization domain is present in cells, its cognate binding proteins can thus dimerize with each other, accelerating certain pathway. Membrane Rudder device mentioned before required signal induced dimer, but in transcription signal Membrane Rudder system, we can simply control the assembling of enzymes through transcription signal.
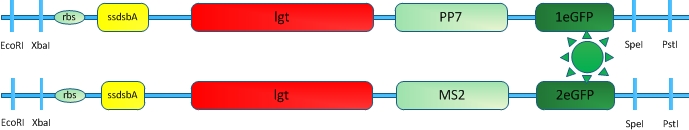 Fig.11 :Fusion protein constructed to test the usability of RNA induced dimerization in our system We incorporated RNA molecule with PP7 and MS2 aptamer domains that bind PP7 and MS2 aptamer binding proteins. (Delebecque, Lindner et al. 2011)
To prove the usability of RNA induced dimerization in our system, we conducted fluorescence complementation assay.
Result
Chemical Signal
There are many ligand induced dimer in organisms. Chemical signal like glutamate receptor(Kunishima, Shimada et al. 2000), ErbB family are ligand-induced dimers(Lemmon 2009) and estrogen receptor which forms dimer when estrogen is present.(Kumar and Chambon 1988
Recruiting those signal induced dimers could easily broaden the application field of our system. What we offer is just a universal tool which can be modified for different use.
|