Team:SEU A/Project
From 2012.igem.org
(Difference between revisions)
(→Overall project) |
|||
| (47 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | |||
| - | |||
<html> | <html> | ||
| - | < | + | <head> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | <meta charset="utf-8"> |
| + | <meta name="viewport" content="width=device-width, initial-scale=1.0"> | ||
| + | <meta name="description" content="Image Wall with jQuery and CSS3" /> | ||
| + | <meta name="keywords" content="jquery, css3, images, wall, thumbnails, slide, animate"/> | ||
| + | <meta name="author" content="Codrops" /> | ||
| + | <title>iGEM 2012 SEU_A Project</title> | ||
| - | + | <link href='http://fonts.googleapis.com/css?family=PT+Sans+Narrow' rel='stylesheet' type='text/css' /> | |
| - | + | <link href='http://fonts.googleapis.com/css?family=Wire+One' rel='stylesheet' type='text/css' /> | |
| - | + | <link href="http://fonts.googleapis.com/css?family=Arvo" rel="stylesheet" type="text/css" /> | |
| - | + | <link rel="stylesheet" href="https://2012.igem.org/Team:SEU_A/main.css?action=raw&ctype=text/css" type="text/css"> | |
| - | + | <link rel="stylesheet" href="https://2012.igem.org/Team:SEU_A/top.css?action=raw&ctype=text/css" type="text/css"> | |
| - | + | <link rel="stylesheet" href="https://2012.igem.org/Team:SEU_A/clear.css?action=raw&ctype=text/css" type="text/css"> | |
| - | + | <link rel="stylesheet" href="https://2012.igem.org/Team:SEU_A/footer.css?action=raw&ctype=text/css" type="text/css"> | |
| - | + | <link rel="stylesheet" href="https://2012.igem.org/Team:SEU_A/navcontainer.css?action=raw&ctype=text/css" type="text/css"> | |
| - | + | <link rel="stylesheet" href="https://2012.igem.org/Team:SEU_A/attribution.css?action=raw&ctype=text/css" type="text/css"> | |
| - | + | <link rel="stylesheet" href="https://2012.igem.org/Team:SEU_A/photowall.css?action=raw&ctype=text/css" type="text/css"> | |
| - | + | <link rel="stylesheet" href="https://2012.igem.org/Team:SEU_A/photowallreset.css?action=raw&ctype=text/css" type="text/css"> | |
| + | <link rel="stylesheet" href="https://2012.igem.org/Team:SEU_A/safety.css?action=raw&ctype=text/css" type="text/css"> | ||
| + | <script src="js/jquery-1.7.1.min.js"></script> | ||
| + | <script src="js/jquery.flexslider-min.js"></script> | ||
| + | <script type="text/javascript" src="http://https://2012.igem.org/Team:SEU_A/top.jq?action=raw&ctype=text/javascript"></script> | ||
| + | <script type="text/javascript" src="http://https://2012.igem.org/Team:SEU_A/top.js?action=raw&ctype=text/javascript"></script> | ||
| - | == | + | <script type="text/javascript" src="http://jqueryjs.googlecode.com/files/jquery-1.3.2.js"></script> |
| + | <script type="text/javascript" src="https://2012.igem.org/Team:SEU_A/attribution/pic.js?action=raw&ctype=text/javascript"></script> | ||
| - | + | <style type="text/css">h1{ font:bold 18px Helvetica, Arial, sans-sarif; color:#FFF; margin:10px 0 0 0; text-align:center; }</style> | |
| + | <style type="text/css">h2{ font-size:24px; padding:10px; }</style> | ||
| - | + | </head> | |
| - | + | <body> | |
| - | + | <div class="navcontainer"> | |
| + | <ul class="navlist"> | ||
| + | <li><a href="https://2012.igem.org/Team:SEU_A">Home</a></li> | ||
| + | <li id="active"><a href="https://2012.igem.org/Team:SEU_A/Project">Project</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:SEU_A/Modeling">Modeling</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:SEU_A/Experiment">Experiment</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:SEU_A/Safety">Safety</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:SEU_A/Humanpractice">Human Practice</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:SEU_A/Attributions">Attributions</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:SEU_A/Team">About Us</a></li> | ||
| + | <li><a href="https://2012.igem.org">iGEM 2012</a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| - | == | + | <div class="container"> |
| + | <header> | ||
| + | <div class="title1"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/a/af/Seua_title.png"> | ||
| + | </div> | ||
| + | <br class="clearfloat" /> | ||
| + | </header> | ||
| + | </div> | ||
| + | </br></br></br> | ||
| + | <div class="content"> | ||
| + | <header> | ||
| + | <h1>Project</h1> | ||
| + | <span>Find our own unique apple.</span> | ||
| + | </header> | ||
| + | </html>[[File:Seua_poison_apple.png|400px|left|Southeast University Project]]<html> | ||
| + | <table id="toc" class="toc" style="margin:0 0 auto auto; background:none; padding:10px;"> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <div id="toctitle"><h2>Index</h2> </div> | ||
| + | <ul> | ||
| + | <li class="toclevel-1 tocsection-0"><a href="#.A0"><span class="tocnumber">1</span> <span class="toctext">Background</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-0"><a href="#.B0"><span class="tocnumber">2</span> <span class="toctext">The Story - Toxic Apple</span></a></li> | ||
| + | <li class="toclevel-3 tocsection-0"><a href="#.C0"><span class="tocnumber">3</span> <span class="toctext">Five objective parts involved:</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-3 tocsection-1"><a href="#.C1"><span class="tocnumber"> 3.1</span> <span class="toctext">Environment Condition report Genes</span></a></li> | ||
| + | <li class="toclevel-3 tocsection-2"><a href="#.C2"><span class="tocnumber"> 3.2</span> <span class="toctext">Regulator Genes</span></a></li> | ||
| + | <li class="toclevel-3 tocsection-3"><a href="#.C3"><span class="tocnumber"> 3.3</span> <span class="toctext">Sweet Genes</span></a></li> | ||
| + | <li class="toclevel-3 tocsection-4"><a href="#.C4"><span class="tocnumber"> 3.4</span> <span class="toctext">Undamental Conjugation Device</span></a></li> | ||
| + | <li class="toclevel-3 tocsection-5"><a href="#.C5"><span class="tocnumber"> 3.4</span> <span class="toctext">Death Genes</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="toclevel-4 tocsection0"><a href="#.C0"><span class="tocnumber">4</span> <span class="toctext">Bdellovibrio Detector(Dog Eat Dog System)</span></a></li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </br><h2> <span class="mw-headline" id=".A0">1 Background</span></h2></br> | ||
| + | </html>[[File:Seua_project_background.jpg|300px|left|Southeast University Project Background]]<html> | ||
| + | As we all know, since the discovery of penicillin, people gradually rely on various antibiotics to kill bacteria. However, the adaptation ability of bacteria that of responding to these medicines is out of our imagination , the speed of production of resistance to drugs is faster than creating a new effective drug as well. We come across a big dilemma in drug resistance when fight with the bacteria. Consequently, a new and effective method of killing bacteria is in urgent need.</br> | ||
| + | Based on this consideration, we come up with two innovative ways which have great advantage over the traditional one.</br></br> | ||
| + | </html>[[File:Seua_project.jpg|200px|right|Southeast University Project]]<html> | ||
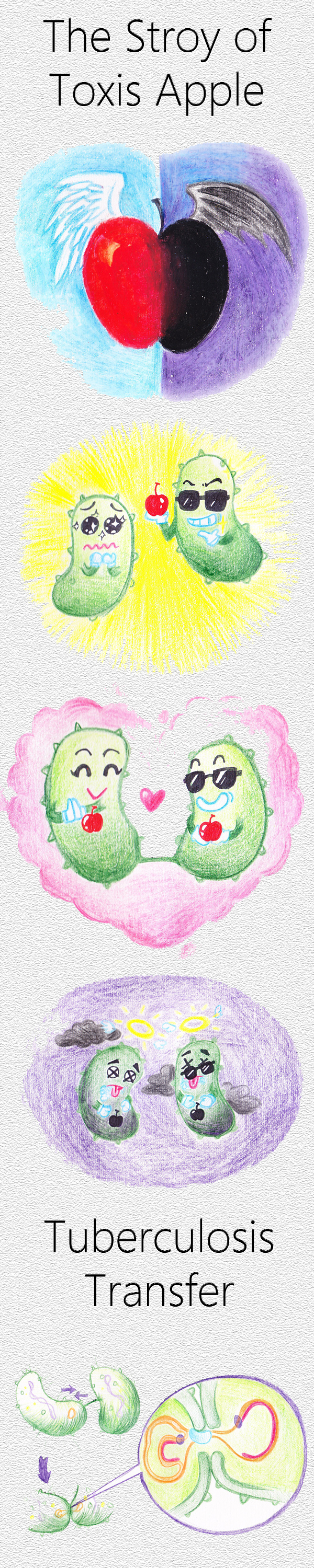
| + | </br><h2> <span class="mw-headline" id=".B0">2 The Story - Toxic Apple</span></h2></br> | ||
| + | The idea is from the bacteria's own selection system and we try to take advantage of it. We intend to construct a system through which bacteria will transfer the death gene by themselves. The question is bacteria won’t keep the gene that play negative role to their survival and will drop the system someday. In order to maintain or even widely spread the suicide system we endow it with the sweet gene that will has positive affection to the multiplication of bacteria. Regularly, the sweet gene expresses to make bacteria glad to accept it, promoting the spread of plasmid from one bacteria to another through conjugative transfer (a process bacteria inherently own) in some degree. Once its account reaches a certain threshold, the dead gene switch will turn on, resulting in the death of the host bacteria. So, function of the whole device can be abstractly described as the toxic apple in fairy tale.</br></br> | ||
| - | === | + | </br><h2> <span class="mw-headline" id=".C0">3 Five Objective Parts Involved</span></h2></br> |
| + | <span class="mw-headline" id=".C1">3.1 Environment Condition report Genes</span></br></br> | ||
| + | This part is aim at report the temperature of the environment.( K873002 is a heat shock promoter that transcribes when the temperature suddenly rises from37℃ to 42℃). Because the sweet genes show obvious benefits to the bacteria under the heat shocking condition, we would like to set a GFP reporter in order to ensure the sweet genes effects.</br></br> | ||
| + | <span class="mw-headline" id=".C2">3.2 Regulator Genes</span></br></br> | ||
| + | This part has regulative function. At the present stage of our project, we wish to improve the conjugation rate by increasing the density of bacteria. So, we use K873001(refer to the flagellar motility of E.coli) and R0084(At low osmolarity activating transcription) to made the bacteria gathering.</br></br> | ||
| + | <span class="mw-headline" id=".C3">3.3 Sweet Gene</span></br></br> | ||
| + | Sweet genes is the part that has positive affection to the multiplication of bacteria to help conceal and spread the death genes. We use molecule chaperone groE(K873000) which is found in many bacteria as the sweet genes cause it is required for proper folding of many proteins.</br></br> | ||
| + | <span class="mw-headline" id=".C4">3.4 Undamental Conjugation Device</span></br></br> | ||
| + | The whole system relays on this part to transfer through conjugation. Among the part, J01003 is the key gene to be mobilized by R plasmid and we just add the I13521(RFP reporter) to test the mobility.</br></br> | ||
| + | <span class="mw-headline" id=".C5">3.5 Death Genes</span></br></br> | ||
| + | We are glad to find and use the ready-made parts that satisfied our requirements. K117010 can produce lysis protein which has the fatal impact on bacteria, meanwhile, the promoter of the part be induced by AI-2 has the detection system to make the suicide process more intelligent. What’s more, it provides the opportunity to set the threshold of the bacteria count.</br></br> | ||
| + | </br><h2> <span class="mw-headline" id=".D0">4 Bdellovibrio Detector(Dog Eat Dog System)</span></h2></br> | ||
| + | The second method derives from the idea of cannibalism, we settle on a Bdellovibrio bacteriovorus strain, who lived in cracking other bacteria. We consider to reconstruct the chemotaxis of Bdellovibrio bacteriovorus,which leads to some sort of special chemotaxis action,and then improves the sterilization efficiency to a certain bacteria. Besides due to the special sterilization of Bdellovibrio bacteriovorus that can decompose the genetic material of the target bacteria, it can diminish the probability of bacteria resistance at the same time.</br></br> | ||
| + | </div> | ||
| + | <div class="footer"> | ||
| + | <a href="https://2012.igem.org/Main_Page/" target="_blank">iGEM 2012</a> | ||
| + | <a href="/wiki/index.php?title=Team:SEU_A/Project&action=edit">Edit</a> | ||
| + | <a href="#top" onclick="window.scrollTo(0,0);return false;">Top</a> | ||
| + | </div> | ||
| - | |||
| + | </br></br></br> | ||
| + | <p align="center"><a href="http://www.seu.edu.cn" target="_blank"><strong>Southeast University</strong></a></p> | ||
| + | <p align="center"><a href="http://bme.seu.edu.cn/" target="_blank">Biomedical Engineer School, SEU </a> | <a href="https://2012.igem.org/" target="_blank"> iGEM 2012</a></p> | ||
| + | <p align="center">Copyright © Southeast University iGEM 2012 Team A, All rights reserved.</p> | ||
| - | + | </body> | |
| - | + | </html> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 21:03, 26 September 2012

Project
Find our own unique apple.
Index |
1 Background
As we all know, since the discovery of penicillin, people gradually rely on various antibiotics to kill bacteria. However, the adaptation ability of bacteria that of responding to these medicines is out of our imagination , the speed of production of resistance to drugs is faster than creating a new effective drug as well. We come across a big dilemma in drug resistance when fight with the bacteria. Consequently, a new and effective method of killing bacteria is in urgent need. Based on this consideration, we come up with two innovative ways which have great advantage over the traditional one.2 The Story - Toxic Apple
The idea is from the bacteria's own selection system and we try to take advantage of it. We intend to construct a system through which bacteria will transfer the death gene by themselves. The question is bacteria won’t keep the gene that play negative role to their survival and will drop the system someday. In order to maintain or even widely spread the suicide system we endow it with the sweet gene that will has positive affection to the multiplication of bacteria. Regularly, the sweet gene expresses to make bacteria glad to accept it, promoting the spread of plasmid from one bacteria to another through conjugative transfer (a process bacteria inherently own) in some degree. Once its account reaches a certain threshold, the dead gene switch will turn on, resulting in the death of the host bacteria. So, function of the whole device can be abstractly described as the toxic apple in fairy tale.3 Five Objective Parts Involved
3.1 Environment Condition report Genes This part is aim at report the temperature of the environment.( K873002 is a heat shock promoter that transcribes when the temperature suddenly rises from37℃ to 42℃). Because the sweet genes show obvious benefits to the bacteria under the heat shocking condition, we would like to set a GFP reporter in order to ensure the sweet genes effects. 3.2 Regulator Genes This part has regulative function. At the present stage of our project, we wish to improve the conjugation rate by increasing the density of bacteria. So, we use K873001(refer to the flagellar motility of E.coli) and R0084(At low osmolarity activating transcription) to made the bacteria gathering. 3.3 Sweet Gene Sweet genes is the part that has positive affection to the multiplication of bacteria to help conceal and spread the death genes. We use molecule chaperone groE(K873000) which is found in many bacteria as the sweet genes cause it is required for proper folding of many proteins. 3.4 Undamental Conjugation Device The whole system relays on this part to transfer through conjugation. Among the part, J01003 is the key gene to be mobilized by R plasmid and we just add the I13521(RFP reporter) to test the mobility. 3.5 Death Genes We are glad to find and use the ready-made parts that satisfied our requirements. K117010 can produce lysis protein which has the fatal impact on bacteria, meanwhile, the promoter of the part be induced by AI-2 has the detection system to make the suicide process more intelligent. What’s more, it provides the opportunity to set the threshold of the bacteria count.4 Bdellovibrio Detector(Dog Eat Dog System)
The second method derives from the idea of cannibalism, we settle on a Bdellovibrio bacteriovorus strain, who lived in cracking other bacteria. We consider to reconstruct the chemotaxis of Bdellovibrio bacteriovorus,which leads to some sort of special chemotaxis action,and then improves the sterilization efficiency to a certain bacteria. Besides due to the special sterilization of Bdellovibrio bacteriovorus that can decompose the genetic material of the target bacteria, it can diminish the probability of bacteria resistance at the same time.Biomedical Engineer School, SEU | iGEM 2012
Copyright © Southeast University iGEM 2012 Team A, All rights reserved.
 "
"