Team:Potsdam Bioware/Lab/Labjournal/July
From 2012.igem.org
AID
2012-07-02
Topic: Overnight culture of CMV and polyA carrying cells
Investigators: Mario, Tom S.
Time: 2012-07-02
AIM: Preparation of wild type AID
Materials:
- LB medium
- Chloramphenicol 25 mg/mL stock solution in 70 % EtOH
- Plasmids: pSB1C3 with CMV; pSB1C3 with Poly-A
Method:
Inoculation of cell sample each in 5 ml LB medium
shaking over night at 37 °C, 300 rpm, approx. 16 hours
Further tasks:
- Miniprep
2012-07-03
Topic: Glycerolstocks, Miniprep and preparative digestion
Investigators: Basia, Tom S., Chris, Mario
Time: 2012-07-03
Aim: Preparation of wildtype AID
Materials:
- Glycerol
- Miniprep Kit
- overnight culture (pSB1C3 with CMV); overnight culture (pSB1C3 with Poly-A)
- CMV: Restriction enzymes (SpeI, PstI); NEB buffer 2
- Poly-A: Restriction enzymes (Poly A: PstI, XbaI); NEB buffer 3
Method:
Glycerol stock: 500 µL Glycerol 99,8 % + 500 µL overnight cultures --> put in -80 °C freezer
Miniprep (both over night culture (pSB1C3 with CMV) and over night culture (pSB1C3 with Poly-A)
preparative digestion: 25 µL DNA + 1 µL of each enzyme + 3 µL NEB (2 or 3) buffer
Results:
DNA - concentrations via nanodrop:
pcDNA5 (AG) = 642,9 ng/µL
pcDNA5 (good) = 729,1 ng/µL
pcDNA5 (bad) = 705,4 ng/µL
pSB1C3 with CMV = 311,9 ng/µL
pSB1C3 with Poly-A = 360,3 ng/µL
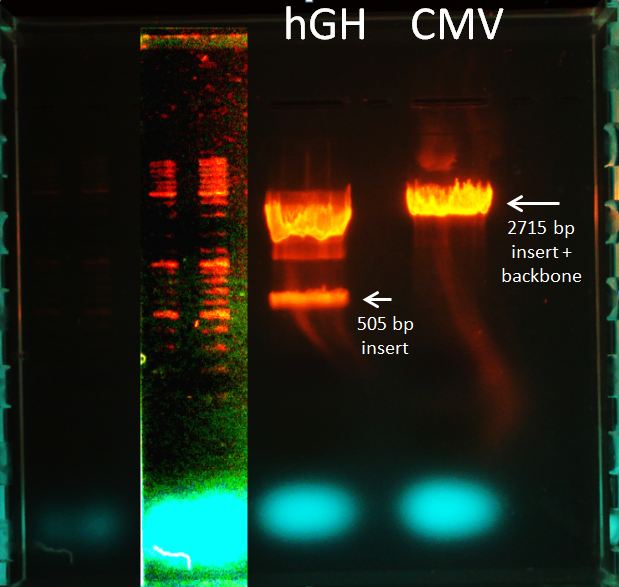
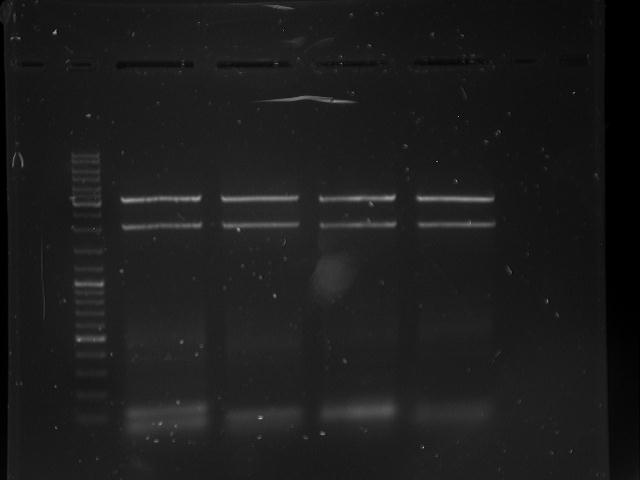
Topic: Separation of cut DNA fragments via gel electrophoresis
Investigators: Chris, Mario
Time: 2012-07-03
Aim Separation of cut DNA fragments via gel electrophoresis
Materials:
gel electrophoresis material
cut samples:
- CMV: Restriction enzymes (SpeI, PstI); NEB buffer 2
- Poly-A: Restriction enzymes (PstI, XbaI); NEB buffer 3
Method:
samples:
- 30 µL CMV cut with SpeI and PstI + 7,5 µL loading dye
- 30 µL polyA + 7,5 µL loading dye
gel electrophoresis conditions:
30 µL of each sample into one big slot
V = 120 V
duration roughly 50 minutes
Results:
Marked fragments were cut out of the gel and transferred into 1,5 mL Eppendorf tubes
Further Tasks:
Gel Extraction
2012-07-04
Gel Extraction of CMV and polyA
Investigators:
Mario, Tom S.
Aim:
Gel Extraction of CMV and polyA
Materials:
centrifuge, Nucleo Spin and PCR clean up - Kit, thermo block, nanodrop
Molecular weight calculator online resource: http://www.encorbio.com/protocols/Nuc-MW.htm
multiplicate with factor 2 when DNA is double stranded
Method:
extract DNA: according to the manual
Results:
DNA-concentrations via nanodrop:
CMV = 106,8 ng/µL -> 63,7 nM (with mass conc. of 1676525,6 Da)
polyA = 15,1 ng/µL -> 48,5 nM (with mass conc. of 311550 Da)
location: -20 °C freezer, topmost drawer
ready DNA for Ligation
Further tasks:
ligation of fragments
Topic: Overnight culture of AID carrying cells
Investigators: Sascha
Time: 2012-07-04
Materials:
LB medium
ampicillin 100 mg/ ml stock solution
glycerol stocks E. coli XL1 blue with plasmids: pSB1C3 with AID
Method:
Inoculation of cell sample in 5 ml LB medium
shaking overnight at 37°C, 300 rpm, approx. 16 hours
Further tasks:
Miniprep
2012-07-05
Topic: Miniprep and preparative digestion
Investigators: Chris
Time: 2012-07-05
Materials:
Miniprep Kit
over night culture (pSB1A3 with AID)
AID: Restriction enzymes (XbaI, PstI); NEB buffer 3
Method:
Miniprep according to the manual
preparative digestion: 25 µL DNA + 1 µL of each enzyme + 3 µL NEB 3 buffer
Results:
DNA - concentrations via nanodrop:
pSB1A3 with AID = 85,5 ng/µL
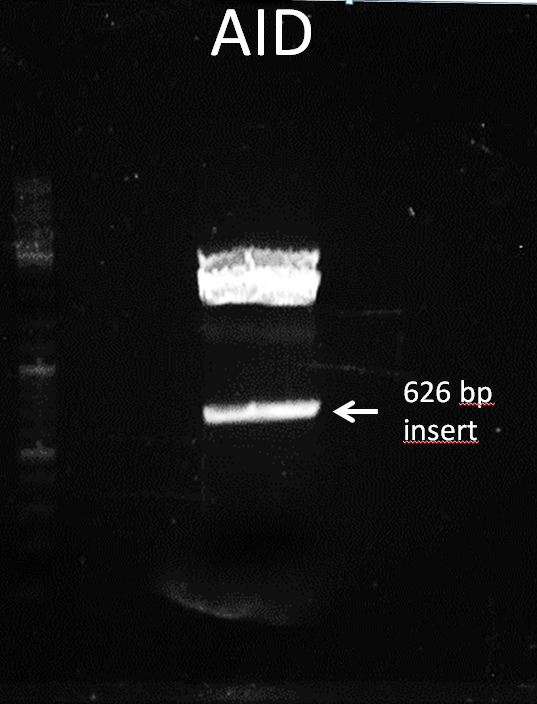
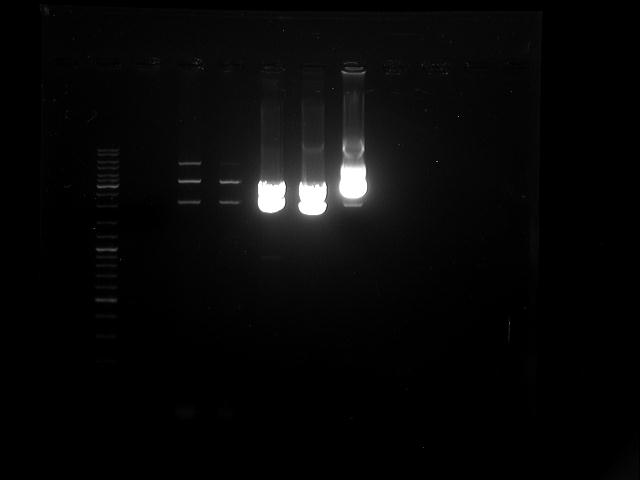
Topic: Separation of cut DNA fragments via gel electrophoresis
Investigators: Mario, Tom S.
Time: 2012-07-05
Aim Separation of cut DNA fragments via gel electrophoresis
Materials:
gel electrophoresis material
cut samples:
AID: Restriction enzymes (XbaI, PstI); NEB buffer 3
Method:
samples:
- 30 µL AID cut with XbaI and PstI + 7,5 µL loading dye
gel electrophoresis conditions:
30 µL of each samples into one big slot converted
V = 120 V
duration roughly 65 minutes
Results:
Marked fragment was cut out of the gel and transferred into 1,5 mL Eppendorf tube
Further Tasks:
Gel extraction
Gel extraction of AID
Investigators:
Mario, Tom S.
Aim:
Gel extraction of AID
Materials:
centrifuge, Nucleo Spin and PCR clean up - Kit, thermo heater, nanodrop
Method:
Gel extraction according to the manual
Results:
DNA-concentrations via nanodrop:
AID = 9,2 ng/µL -> 23,8 nM (with mass conc. of 386864,4 Da)
location: -20 °C freezer, topmost drawer
ready DNA for Ligation
Further tasks:
ligation of fragments
Ligation of CMV (Insert + backbone) and AID (insert)
Investigators:
Mario, Tom S.
Aim:
Ligation of CMV (Insert + backbone) and AID (insert)
Materials:
T4 DNA-Ligase, samples(CMV + AID)
Method:
DNA Fragment ligation: according to the manual
sample preparation:
- 1 µL (CMV Fragment) c=106,8 ng/µL(63,9 nM) -> 6,4 nmol
- 6 µL (AID Fragment) c=9,2 ng/µL(23,8 nM) -> 14,3 nmol
- 1 µL (T4 DNA-Ligase)
- 2 µL (DNase free water)
incubation of sample 1,5 h at 22 °C
Results:
location: -20 °C freezer, topmost drawer
ready DNA Transformation
Further tasks:
Transformation
2012-07-06
Topic: Transformation of ligated sample
Investigators: Mario, Tom S.
Time: 2012-07-06
Materials:
- Bunsen burner, Agar Plate with Chloramphenicol, 37 °C heat block, centrifuge
- ligated sample (compare last step 07-05-2012)
- icebox
- competent E. coli cells (XL 1)
Method:
Transformation via manual
Plate incubation start: 1:30 pm
Results:
grown colonies
Further tasks:
picking clones
2012-07-07
Overnight culture of pSB1C3+CMV+AID carrying cells
Investigators: Chris
Time: 2012-07-07 6pm
Materials:
LB medium, chloramphenicol 25 mg/ ml stock solution, plates with E. coli XL1 blue with plasmids: pSB1C3+CMV+AID
Method: picking clones (2 per plate->Nr.1-6) and inoculation in 5 ml LB medium + 5µl chloramphenicol stock shaking over night at 37°C, 300 rpm, approx. 16 hours
Further tasks:
glycerolstocks & Miniprep
2012-07-08
Topic: Glycerol stocks, Miniprep
Investigators: Basia
Time: 2012-07-08 11:00am
Materials:
Glycerol
Miniprep Kit
6x overnight culture (pSB1C3 with CMV+AID);
Method:
Glycerol stock: 500 µL Glycerol 99,8 % + 500 µL overnight cultures --> put in -80 °C freezer
Miniprep (all 6 over night cultures (pSB1C3 with CMV+AID)
Results:
6 Cryostocks are stored in the igem box in the -80°C freezer and Plasmids are stored in -20°C in the 4th drawer on styrofoam rack
2012-07-09
Topic: Measuring DNA-concentration of plasmids from 2012-07-08
Investigators: Mario, Tom S.
Time: 2012-07-09
Materials:
- Plasmids: pSB1C3 with CMV+AID (samples: 1, 2, 3, 4, 5, 6)
- Nanodrop
- NE-buffer
Method:
2 µL of each DNA-sample onto nanodrop (Ne-buffer blank)
Results:
DNA-concentrations:
1 = 290 ng/µL
2 = 361,2 ng/µL
3 = 316,8 ng/µL
4 = 360,5 ng/µL
5 = 392,5 ng/µL
6 = 390 ng/µL
Further tasks:
- restriction enzyme digestion with XbaI und PstI
Topic: preparative digestion
Investigators: Mario, Tom S.
Time: 2012-07-09
Materials:
- Plasmids: pSB1C3 with CMV+AID (samples: 1, 2, 3, 4, 5, 6)
- Restriction enzymes (XbaI and PstI)
- NE3-buffer
Method:
heat block (37 °C)
sample preparation: each DNA 25 µL + 3 µL NE3-buffer + 1 µL XbaI + 1 µL PstI
incubation of samples for 4 h at 37 °C
Results:
none
Further tasks:
- gel electrophoresis
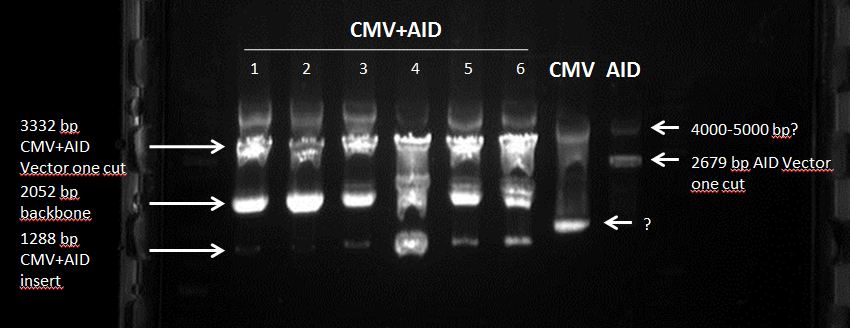
Topic: Separation of cut DNA fragments via gel electrophoresis
Investigators: Mario, Tom S.
Time: 2012-07-09
Aim Separation of cut DNA fragments via gel electrophoresis
Materials:
gel electrophoresis material
cut samples:
AID: Restriction enzymes (XbaI, PstI); NEB buffer 3
CMV: Restriction enzymes (XbaI, PstI); NEB buffer 3
AID+CMV: Restriction enzymes (XbaI, PstI); NEB buffer 3
Method:
samples:
- 10 µL AID cut with XbaI and PstI + 2,5 µL loading dye
gel electrophoresis conditions:
10 µL of each sample into one big well
V = 120 V
duration roughly 95 minutes
Results:
Further Tasks:
overnight culture with AID+CMV sample 1, 2 and 3
Topic: Overnight culture of AID+CMV carrying cells
Investigators: Basia, Tom S.
Time: 2012-07-09, 17:30
Materials:
LB medium
chloramphenicol 25 mg/ ml stock solution
glycerol stocks E. coli XL1 blue with Plasmids: pSB1C3 with AID+CMV of sample 1,2 and 3,
Method:
Inoculation of cell samples in 3 ml LB medium
shaking over night at 37°C, 300 rpm
Further tasks:
Miniprep, preparative digestion, Plasmid ligation with polyA
2012-07-10
Topic: Miniprep of CMV+AID carrying plasmids
Investigators: Tom S., Mario
Time: 2012-07-10
Materials:
- samples(CMV + AID: 1, 2, 3) - for detailed info check lab day 2012-07-09
- Miniprep Kit
- overnight culture (pSB1C3 with AID + CMV: samples 1, 2, 3,)
Method:
Plasmid isolation via Kit (check manual)
concentration measurement via nanodrop (2 µL sample)
Results:
DNA - concentrations via nanodrop:
AID + CMV (1) = 303,2 ng/µL
AID + CMV (2) = 366,6 ng/µL
AID + CMV (3) = 378,5 ng/µL
Further Tasks:
preparative digestion (use sample #3; samples 1 and 2 for back up in -20 °C freezer topmost drawer)
fragment cut
2012-07-11
Topic: preparative digestion
Investigators: Tom S.
Time: 2012-07-11 09:00
Materials:
- pSB1C3 Vector with CMV+AID
- Restriction enzymes (SpeI, PstI); Fast Digest Green Buffer
Method:
preparative digestion: 25 µL DNA + 1 µL of each enzyme + 3 µL Fast Digest Green Buffer (incubation for 2 h)
further tasks:
Gel electrophoresis
Topic: Gel electrophoresis of cut pSB1C3 (CMV + AID) fragments
Investigators: Tom S.
Time: 2012-07-11 11:00
Materials:
- cut sample (CMV + AID, PstI + SpeI)
- Gel electrophoresis material
Method:
sample preparation: none
loading into wll: 30 µL
duration: 70 minutes
Results:
one band
Further Tasks:
Gel extraction
Topic: Gel extraction and measurement of DNA concentration
Investigators: Chris, Mario
Time: 2012-07-11 13:00 - 14:00
Materials:
- Analytic Jena gel extraction kit
- measurement of DNA concentration via nanodrop
Method:
Gel extraction via manual
Results:
DNA concentration via nanodrop: 98.7 ng/µL
-->2051640 Da (2051,64 kDa)--> c=48.1 nM
Further Tasks:
Ligation of fragment with Poly-A
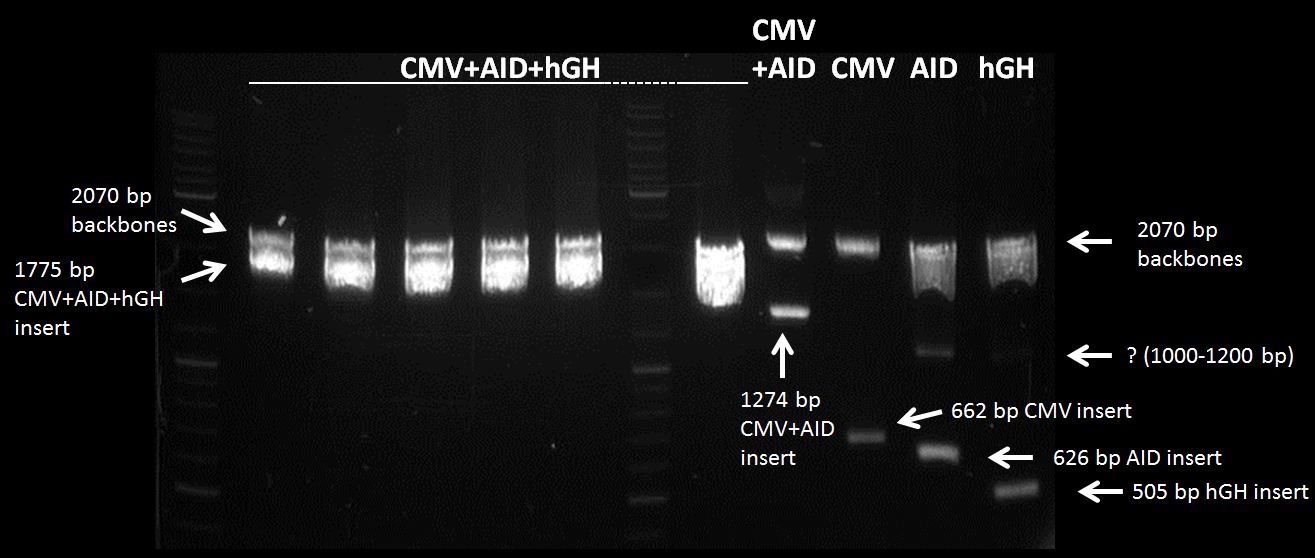
Topic: Ligation CMV+AID in pSB1C3 (cut:Spe1 and Pst1) with hGH-polyA (cut:Xba1 and Pst1)
Investigators: Chris, Mario
Time: 2012-07-11 16:45 - 17:30
Materials:
digested fragments: CMV+AID in pSB1C3 (cut:Spe1 and Pst1) c=48.1 nM , hGH-polyA (cut:Xba1 and Pst1) c= 15,1 ng/µL -> 48,5 nM
Method:
mix 1µL CMV+AID in pSB1C3 (cut:Spe1 Pst1) c=48.1 nM, 3µL hGH-polyA (cut:Xba1 Pst1) c=48.5 nM, 1µl T4 Ligase, 1µ 10x Buffer,4 µL H20
incubate 1.5 h
Results:
not visible
Further Tasks:
Transformation
Topic: Transformation of XL1 Blue with CMV+AID+hGH-polyA in pSB1C3
Investigators: Chris
Time: 2012-07-11 finished:18 Uhr
Materials:
LB medium, E. coli XL1 Blue, ligation product (CMV+AID+hGH-polyA), agar-LB-paltes with 1:1000 chloramphenicol
Method:
transformation - standard operating procedures
Results:
two plates with transformed E. coli (CMV+AID+polyA)
Further Tasks:
picking colonies & inoculate 5 ml overnight culture
2012-07-12
Overnight culture of pSB1C3 CMV+AID+hGH-polyA, CMV+AID, CMV, hGH-polyA and pSB1A3 AID carrying cells
Investigators: Tom S.
Time: 2012-07-12 6pm
Materials:
LB medium, chloramphenicol 25 mg/ ml stock solution, plates with E. coli XL1 blue with plasmids: pSB1C3+CMV+AID, glycerol stocks: pSB1C3 with AID+CMV, CMV, hGH-polyA and pSB1A3 with AID
Method: picking clones(3 per plate->Nr.1-6) and inoculation in 5 ml LB medium + 5µl chloramphenicol stock shaking over night at 37°C, 300 rpm, approx. 16 hours, samples from glycerolstocks in 3 ml LB + 3 µL chloramphenicol or 3µL Amp
Further tasks:
glycerol stocks & Miniprep
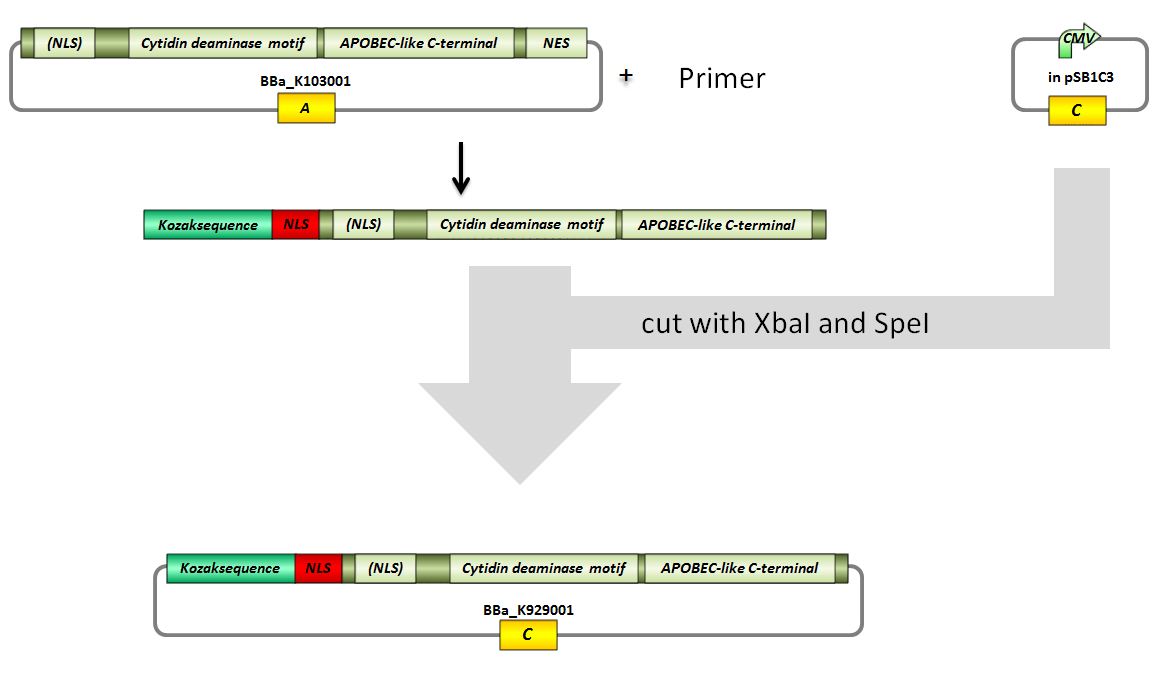
Topic: Planing BBa_K929001
Investigators: Tom S., Chris, Basia, Rico, Mario, Kevin
Aim: planing how to digest and ligate the vectors for BBa_K929001
Material: Genious
Results:
pSB1C3 with CMV -> (cut with SpeI and XbaI) 2072 bp (pSB1C3 backbone) + 662 bp (rest)
PCR-amplificate -> (cut with SpeI and XbaI) 597 bp (modified AID insert)
Further tasks:
design and ordering of primers, practical part
Primer design and ordering for BBa_K929001
Investigators: Tom S., Rico
Time: 2012-07-12 7pm
Primer (forward) with XbaI recognition site, kozak consensus sequence, NLS:
ATCTAGAGCCGCCACCATGGGACCCAAGAAGAGGAAGGTGATGGACAGCCTCTTGATGAACCGGAGG
Primer (reverse, complement)with AgeI and SpeI recognition site:
CCACTAGTATTAACCGGTGGGCAAAAGGATGCGCCGAAGC
2012-07-13
Mini Prep of WT Plasmids, nanodrop
Investigators: Tom S., Mario
Time: 2012-07-13 10am
Materials:
Miniprep Kit
Overnight culture of AID-WT tranfected E. coli strains
Method: Kit via manual
Results:
DNA-concentrations via nanodrop:
WT-AID 1: 387,2 ng/µL
WT-AID 2: 453,0 ng/µL
WT-AID 3: 415,8 ng/µL
WT-AID 4: 445,5 ng/µL
WT-AID 5: 474,1 ng/µL
WT-AID 6: 645,1 ng/µL
AID: 393,5 ng/µL
CMV: 188,0 ng/µL
CMV+AID 221,9 ng/µL
hGH 318,2 ng/µL
Further tasks:
digestion and gelelectrophoresis
Topic: preparative digestion
Investigators: Tom S.
Time: 2012-07-11 09:00
Materials:
- pSB1C3 Vectors with CMV+AID+hGH-polyA, AID, CMV, hGH, CMV+AID
- Restriction enzymes (SpeI, PstI); Fast Digest Green Buffer
Method:
preparative digestion: 25 µL DNA + 1 µL of each enzyme + 3 µL Fast Digest Green Buffer (incubation for 2 h)
further tasks:
Gel electrophoresis
2012-07-16
Gel electrophoresis of cut ligation samples (WT AID - CMV+AID+hGH-polyA)
Investigators: Tom S.
Time: 2012-07-16;
Materials:
gel electrophoresis equipment
samples
Method:
loading wells with 10 µL of each cut sample (ca. 600-800 ng DNA per sample)
gel ectrophoresis standard operating procedure
Results:
Further tasks:
sequencing
2012-07-23
Topic: PCR of AID+NLS+Kozak sequence
Investigators:
Basia, Tom S.
Aim:
- amplification of the AID with inserted Kozak sequence and NLS sequence via PCR
Materials:
- Phusion, template (AID insert), Primers designed by Tom S. and Rico on 12.07.2012, dNTPs, Polymerase)
- PCR clean-up kit
Method:
- polymerase chain reaction
Mastermix
| reagent | volume [µL] |
| HF Phusion buffer 5x | 10 |
| dNTPs | 1 |
| Primer (Forward) | 1,25 |
| Primer (Reverse) | 1,25 |
| DNA (Plasmid) | 1,0 |
| Phusion Polymerase | 0,5 |
| water | 35,0 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| denaturation | 98 | 30 | 1 |
| denaturation | 98 | 5 | 17 |
| annealing + elongation | 72 | 45 | 17 |
| denaturation | 98 | 5 | 17 |
| elongation | 72 | 25 | 17 |
| final elongation | 72 | 600 | 1 |
| cooling | 4 | ∞ | 1 |
Results:
125ng/µl - 1st sample, 135ng/µl 2nd sample
Further tasks:
- digestion + agarose gel electrophoresis
Primer design and ordering for sequencing BBa_K929001 and BBa_K929003
Investigators: Tom S., Rico
Time: 2012-07-23
Primer bind on pSB1C3-vector (left next to backbone-prefix):
GGCGTATCACGAGGCAG
Primer (reverse, complement) bind on pSB1C3-vector (right next to backbone-suffix):
CGAGTCAGTGAGCGAGG
2012-07-25
Topic: preparative digestion
Investigators: Tom S.
Time: 2012-07-25 08:30
Materials:
pSB1C3 Vector with CMV
2 PCR-products - AID without NES, with NLS+Kozak Sequence (theoretically the same) (SpeI, XbaI; Fast Digest); Fast Digest Green Buffer
Method:
preparative digestion: 25 µL DNA + 1 µL of each enzyme + 3 µL Fast Digest Green Buffer (incubation for 2 h)-> for the pSB1C3 backbone
preparative digestion: 18 µL DNA + 1 µL of each enzyme + 2 µL Fast Digest Green Buffer (incubation for 2 h)-> for the PCR-products
Topic: Separation of cut DNA fragments via gel electrophoresis
Investigators: Tom S.
Time: 2012-07-25
Aim Separation of cut DNA fragments via gel electrophoresis
Materials:
gel electrophoresis material
Method:
cut samples:
- pSB1C3 backbone: Restriction enzymes (XbaI, SpeI; Fast Digest); Fast Digest buffer
- PCR-products (AID without NES, with NLS+Kozak Sequence): Restriction enzymes (XbaI, SpeI; Fast Digest); Fast Digest buffer
wells loaded with 30 or 22 µL of digested samples via gel electrophoresis - standard operating procedure
gel electrophoresis conditions:
V = 120 V
duration roughly 50 minutes
Results:
Marked fragment was cut out of the gel and transferred into 1,5 mL Eppendorf tube
Further Tasks:
Gel extraction
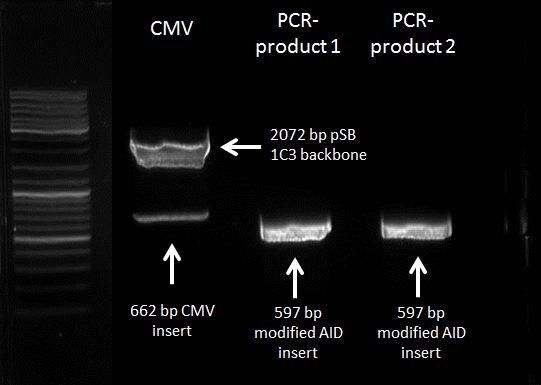
Gel extraction of pSB1C3 backbone and modified AID insert (AID without NES, with NLS+Kozak Sequence)
Investigators:
Tom S.
Aim:
Gel extraction of pSB1C3 backbone and modified AID insert
Materials:
centrifuge, Nucleo Spin and PCR clean up - Kit, thermo heater, nanodrop
Method:
DNA extraction: according to the manual
Results:
DNA-concentrations via nanodrop:
pSB1C3 backbone = 83,6 ng/µL -> 65,7 nM (with mass conc. of 1273,3 kDa)
PCR-product 1 = 74,0 ng/µL -> 201,5 nM (with mass conc. of 367,18 kDa)
PCR-product 1 = 77,5 ng/µL -> 211,1 nM (with mass conc. of 367,18 kDa)
location: -20 °C freezer, topmost drawer
ready DNA for ligation
Further tasks:
ligation of fragments
Ligation of PCR-product (AID without NES, with NLS+Kozak Sequence) and pSB1C3 backbone
Investigators:
Tom S.
Aim:
Ligation of PCR-product and pSB1C3 backbone
Materials:
T4 DNA-Ligase, samples(PCR-product 1 and 2 + pSB1C3 backbone)-> PCR products 1 and 2 are theoretically the same
Method:
DNA Fragment ligation: according to the manual
sample preparation:
- 2 µL (PCR-product) c=75,8 ng/µL(206,4 nM)-> 41,3 nM
- 2 µL (pSB1C3 backbone) c=83,6 ng/µL(65,7 nM) -> 13,2 nM
- 1 µL (T4 DNA-Ligase)
- 1 µL 10x T4 DNA Ligase Buffer
- 4 µL (DNase free water)
incubation of sample 1,5 h at 22°C
Results:
samples ligated
location: -20 °C freezer, topmost drawer
ready DNA for transformation
Further tasks:
Transformation
Topic: Transformation of ligated sample
Investigators: Tom S.
Time: 2012-07-25
Materials:
- Bunsen Burner, Agar Plate with Chloramphenicol, 37 °C heater, centrifuge
- ligated sample (compare last step 25-07-2012)
- icebox
- competent E. coli cells (XL 1)
Method:
Transformation via manual
Plate incubation start: 5:00 pm
Results:
ready for growing mutants to pick clones
Further tasks:
picking clones
2012-07-26
picking clones & inoculation
Investigators: Sascha
Time: 2012-07-27 6 pm
Materials:
LB medium, chloramphenicol 25 mg/ ml stock solution, plates with E. coli XL1 blue with plasmids: pSB1C3+pcr-products(AID with NLS,without NES+Kozak sequence), glycerol stocks: pSB1C3 with CMV
Method: picking clones(3 per plate) and inoculation in 5 ml LB medium + 5µl chloramphenicol stock shaking over night at 37°C, 300 rpm, approx. 16 hours, samples of glycerol stocks in 5 ml LB + 5 µL chloramphenicol
Further tasks:
glycerolstocks & Miniprep
2012-07-27
Miniprep
Investigators: Tom S.
Time:
2012-07-03 8 am
Materials:
Glycerol, Miniprep Kit, over night culture (pSB1C3 with CMV); overnight culture (pSB1C3 with modified AID)
Method:
Glycerol stock: 300 µL Glycerol 99,8 % + 700 µL over night cultures --> put in -80 °C freezer
Miniprep (both overnight culture (pSB1C3 with CMV) and overnight cultures (pSB1C3 with PCR 1 colony 1-3 and PCR 2 colony 1-3)
Results:
DNA - concentrations via nanodrop:
PCR1 C1= 163.7 ng/µL
PCR1 C2= 154.7 ng/µL
PCR1 C3= 165.5 ng/µL
PCR2 C1= 117.1 ng/µL
PCR2 C2= 164.7 ng/µL
PCR2 C3= 94,4 ng/µL
CMV= 144.2 ng/µL
Further tasks:
preparative digestion
Topic: preparative digestion
Investigators:Chris
Time: 2012-07-27 11:30
Materials:
pSB1C3 Vector with CMV (SpeI, PstI; Fast Digest); Fast Digest Green Buffer
PCR 1 C 3 and PCR 2 C 2 in pSB1C3 vector(PstI, XbaI; Fast Digest); Fast Digest Green Buffer
Method:
- preparative digestion: 25 µL DNA + 1 µL of each enzyme + 3 µL Fast Digest Green Buffer (incubation for 2,5 h)
Topic: Separation of cut DNA fragments via gel electrophoresis
Investigators: Tom S.
Time: 2012-07-27
Aim: Separation of cut DNA fragments via gel electrophoresis
Materials:
gel electrophoresis material
cut samples:
- digested CMV: Restriction enzymes (SpeI, PstI; Fast Digest); Fast Digest buffer
- digested PCR-products: Restriction enzymes (PstI, XbaI; Fast Digest); Fast Digest buffer
Method:
samples:
loading wells with 30µl of digested samples via gel electrophoresis, standard operating procedure
gel electrophoresis conditions:
30 µL of each samples into one big well
V = 120 V
duration 72 minutes
Results:
Marked fragments were cut out of the gel and transferred into 1,5 mL Eppendorf tube for gel extraction
Further Tasks:
Gel extraction
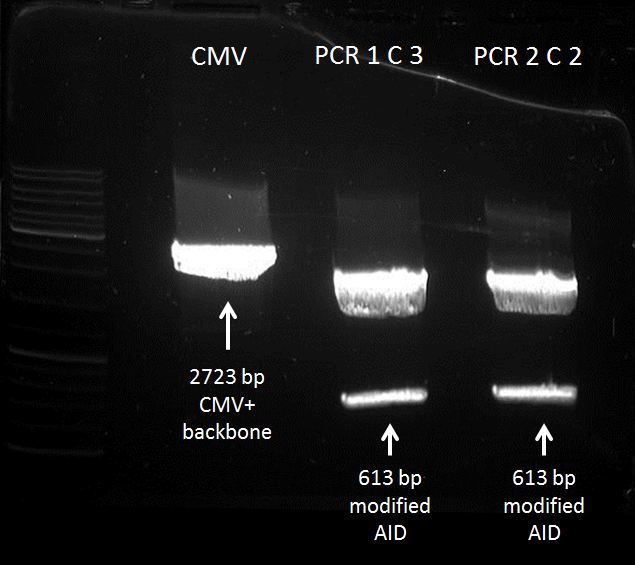
Gel extraction of digested CMV+backbone and PCR-products (AID without NES, with NLS+Kozak sequence) insert
Investigators:
Tom S.
Aim:
Gel extraction of digested CMV+backbone and PCR-products insert
Materials:
centrifuge, Nucleo Spin and PCR clean up - Kit, heat block, nanodrop
Method:
extraction of DNA: according to the manual
Results:
DNA-concentrations via nanodrop:
CMV+backbone = 85,6 ng/µL -> 50,9 nM (with mass conc. of 1681,3 kDa)
PCR 1 C 3 = 33,7 ng/µL -> 89,1 nM (with mass conc. of 378,3 kDa)
PCR 2 C 2 = 29,4 ng/µL -> 77,7 nM (with mass conc. of 378,3 kDa)
location: -20 °C freezer, topmost drawer
ready DNA for Ligation
Further tasks:
ligation of fragments
2012-07-28
Ligation of PCR-products(AID without NES, with NLS+Kozak sequence) and CMV+backbone
Investigators:
Basia
Aim:
Ligation of PCR-products and CMV+backbone
Materials:
T4 DNA-Ligase, samples(PCR 1 C 3 or PCR 2 C 2 and CMV+backbone)
Method:
DNA fragment ligation: according to the manual
sample preparation:
- 4 µL PCR 1 C3 c=33,7 ng/µL(89,1 nM)-> 35,6 nM; (for the other sample 4µL PCR 2 C 2 c=29,4 ng/µL(77,7 nM)-> 31,1 nM
- 2 µL (CMV+backbone) c=85,6 ng/µL(50,9 nM) -> 10,2 nM
- 1 µL (T4 DNA-Ligase)
- 1 µL 10x T4 DNA Ligase Buffer
- 2 µL (DNase free water)
incubation of sample 1,5 h at 22°C
Results:
location: -20°C freezer, topmost drawer
ready DNA Transformation
Further tasks:
Transformation
Topic: Transformation of ligated sample
Investigators: Basia
Time: 2012-07-28 12:30
Materials:
- Bunsen Burner, Agar Plate with Chloramphenicol, 37°C heat block, centrifuge
- ligase sample (from last step 28.07.2012)
- icebox
- competent E. coli cells (XL 1 Blue)
Method:
Transformation according to the manual
Plate incubation start: 14:30 pm
Results:
ready mutants to pick clones
Further tasks:
picking clones
2012-07-29
Overnight culture of E. coli containing plasmids with PCR products (AID without NES, with NLS+Kozak sequence) and CMV promoter
Investigators: Basia
Time: 2012-07-29 7 pm
Materials:
LB medium, chloramphenicol 25 mg/ ml stock solution, plates with E. coli XL1 blue with plasmids: CMV+PCR1C3 and CMV+PCR2C2
Method: picking clones(2 per plate) and inoculation in 5 ml LB medium + 5µl chloramphenicol stock shaking over night at 37°C, 300 rpm, approx. 16 hours
Further tasks:
glycerol stocks & Miniprep
2012-07-30
Topic: Transformation of BBa_E0040 (wild-type GFP) from Distribution Plate 1 Kit 2012
Investigators: Chris
Time: 2012-07-30 10:30
Materials:
Bunsen Burner, Agar Plate with ampicillin, icebox, competent E. coli cells (XL 1 Blue)
Method:
Transformation according to the manual
Plate incubation start: 13:30 pm
Results:
ready mutants to pick clones
Further tasks:
picking clones
Glycerol stocks & miniprep of CMV+AID without NES, with NLS+Kozak sequence containing plasmids
Investigators: Tom S.
Time:
2012-07-03 8:30 am
Materials:
Glycerol, Miniprep Kit, overnight cultures (CMV+PCR1C3C1-2; CMV+PCR2C2C1-2)
Method:
Glycerol stock: 300 µL Glycerol 99,8 % + 700 µL overnight cultures --> put in -80 °C freezer
Miniprep overnight cultures (CMV+PCR1C3C1-2; CMV+PCR2C2C1-2) procedure according to the manual
Results:
DNA concentrations via nanodrop:
PCR1C3 C1= 389,5 ng/µL
PCR1C3 C2= 394,3 ng/µL
PCR2C2 C1= 409,8 ng/µL
PCR2C2 C2= 383,4 ng/µL
Further tasks:
preparative digestion with PCR1C3 C2 and PCR2C2 C1
Topic: preparative digestion
Investigators:Chris
Time: 2012-07-30 11:00
Materials:
pSB1C3 Vector with hGH-PolyA (XbaI, PstI; Fast Digest); Fast Digest Green Buffer
PCR1C3 C2 and PCR2C2 C1 in pSB1C3 vector - AID without NES, with NLS+Kozak sequence+CMV(PstI, SpeI; Fast Digest); Fast Digest Green Buffer
Method:
preparative digestion: 25 µL DNA + 1 µL of each enzyme + 3 µL Fast Digest Green Buffer (incubation for 2 h)
Topic: Separation of cut DNA fragments via gel electrophoresis & Gel extraction
Investigators: Chris
Time: 2012-07-30
Aim: Separation of cut DNA fragments via gel electrophoresis
Materials:
gel electrophoresis material
cut samples:
digested hGH-PolyA: Restriction enzymes (XbaI, PstI; Fast Digest); Fast Digest buffer
digested PCR-products: Restriction enzymes (PstI, SpeI; Fast Digest); Fast Digest buffer
Method:
gel ectrophoresis - standard operating procedure
gel electrophoresis conditions:
30 µL of each samples into one big slot
V = 120 V
duration roughly 60 minutes
Results:
Marked fragments were cut out of the gel and transferred into 1,5 mL Eppendorf tubes
Gel Extraction:
final concentrations:
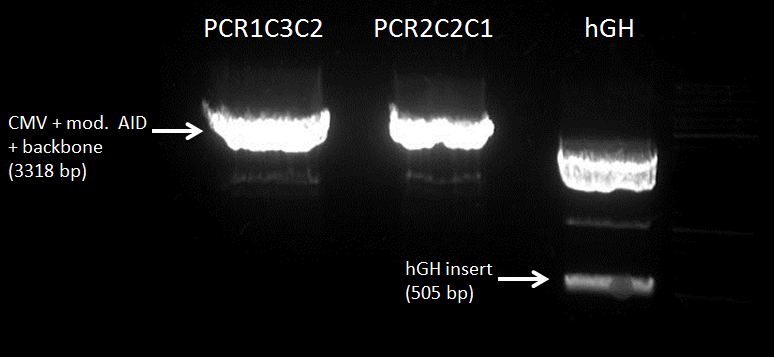
CMV+PCR1C3C2(3318 nt, MW=2048.48 kDa) : 188.1 ng/µl (91.8 nM)
CMV+PCR2C2C1 : 182.2 ng/µl (88.9 nM)
hGH-polyA (495 nt, MW=305.4 kDa): 26.1 ng/µl (85 nM)
Further Tasks:
Ligation, transformation
2012-07-31
Topic: Planing BBa_K929003
Investigators: Tom S., Chris, Basia, Rico, Mario, Kevin
Aim: planing how to digest and ligate the vectors for BBa_K929003
Material: Genious
Results:
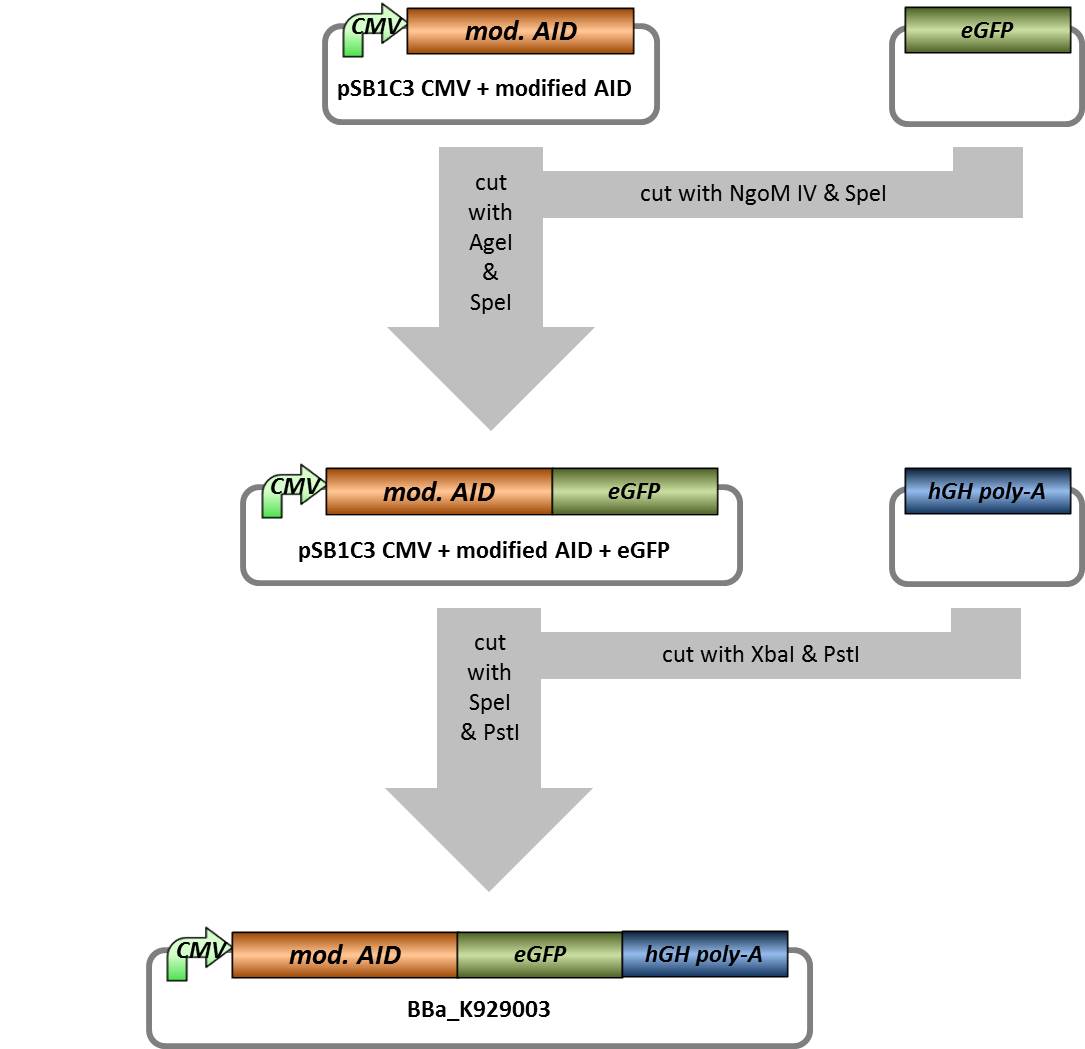
- pSB1C3 with CMV+ mod. AID (without NES, with NLS) -> cut with AgeI and SpeI, 3322 bp - pSB1C3 backbone with CMV + mod. AID, 14 bp - rest
- eGFP -> cut with SpeI and NgoMIV, 731 bp - eGFP insert, 2074 bp - pSB1C3 backbone
- pSB1C3 with CMV+ mod. AID + eGFP -> cut with Spe1 and Pst1, 4035 bp - pSB1C3 backbone with CMV + mod. AID+eGFP, 18 bp - rest
- hGH-polyA -> cut with Xba1 and Pst1, 505 bp - hGH-polyA insert, 2053 - pSB1C3 backbone
Further tasks:
- practical part
inoculation of GFP in pSB1A2 (BBa_E0040) & eGFP (from Sven eGFP in pSB1C3 in RFC 25-like [http://partsregistry.org/Part:BBa_K404316 BBa_K404316]
Investigators: Tom S., Chris
Time: 2012-07-31 3pm
Materials:
LB medium, chloramphenicol 25 mg/ ml stock solution, ampicillin stock solution, plates with E. coli XL1 blue with plasmid: GFP in pSB1A2 (BBa_E0040), glycerol stock from Sven: eGFP in pSB1C3 in RFC 25
Method: picking clones (1 per plate from 2 plates) GFP in pSB1A2 (BBa_E0040) and inoculation in 5 ml LB medium + 5µl ampicillin stock, 2 times inoculation of glycerol stock (eGFP in pSB1C3 in RFC 25) in 5mL LB medium + 5µL chloramphenicol stock, shaking over night at 37°C, 300 rpm, approx. 16 hours
Further tasks:
glycerol stocks & Miniprep
preparation of WT-AID & modified AID (without NES, with NLS) in pSB1C3 for sequencing & send to GATC
Investigators: Tom S., Chris
Time: 2012-07-31 7pm
Materials:
20 µl primer c=10µM for each sequencing, 20 µl sample with concentration around 70 ng/µl
Method: mark the samples with bar code stickers & order the sequencing from GATC, put the cups into the orange boxes
samples:
| sample | barcode | primer | sample | barcode | primer |
| WT AID clone 1 (Cmv+WtAID+PolyA in pSB1C3) | CC0697 | forward pSB1C3 | WT AID clone 1 | CC0709 | reverse pSB1C3 |
| WT AID clone 2 | CC0698 | forward pSB1C3 | WT AID clone 2 | CC0710 | reverse pSB1C3 |
| WT AID clone 3 | CC0699 | forward pSB1C3 | WT AID clone 3 | CC0711 | reverse pSB1C3 |
| WT AID clone 4 | CC0700 | forward pSB1C3 | WT AID clone 4 | CC0712 | reverse pSB1C3 |
| WT AID clone 5 | CC0701 | forward pSB1C3 | WT AID clone 5 | CC0713 | reverse pSB1C3 |
| WT AID clone 6 | CC0702 | forward pSB1C3 | WT AID clone 6 | CC0714 | reverse pSB1C3 |
| PCR1 C1 (modified AID- Kozak sequence+NLS+AID without NES+Age1-restriction site in pSB1C3) | CC0703 | forward pSB1C3 | PCR1 C1 | CC0715 | reverse pSB1C3 |
| PCR1 C2 | CC0704 | forward pSB1C3 | PCR1 C2 | CC0716 | reverse pSB1C3 |
| PCR1 C3 | CC0705 | forward pSB1C3 | PCR1 C3 | CC0717 | reverse pSB1C3 |
| PCR2 C1 | CC0706 | forward pSB1C3 | PCR2 C1 | CC0718 | reverse pSB1C3 |
| PCR2 C2 | CC0707 | forward pSB1C3 | PCR2 C2 | CC0719 | reverse pSB1C3 |
| PCR2 C3 | CC0708 | forward pSB1C3 | PCR2 C3 | CC0720 | reverse pSB1C3 |
Further tasks:
alignment
Antikörper
2012-07-06
Sequencing of pcDNA5-FRT and pOG44 by GATC
Investigators: Sascha
Aim: Sequencing of invitrogen vectors pcDNA5-FRT and pOG44
Materials:
- Geneious
- Lablife Sequences of pcDNA5-FRT and pOG44
- GATC
Method: forward sequencing using GATC-CMV-forward-primer
- pcdna5frt_cmv_forward-CMV-F
- pog44_cmv_forward-CMV-F
Further tasks: checking sequences
Primer-design for assembly-PCR, generating geneconstruct: scFv-TEV-TMD-EYFP
Investigators: Sascha
Aim: Design of primer for assembly-pcr of geneconstruct scFv-TEV-TMD-EYFP
Materials:
- Lablife Sequences of pcDNA5-FRT and pOG44
- Databases
- Oligocalc
- Wordpad
Method:
- P1_Signalp_N-term
- P2_Signalp_C-term
- P3_TMD-N-term
- P4_TMD-C-term/N-YFP
- P5_YFP-N-term/TMD-C-term
- P6_YFP-C-term
- P7_Gesamtanfang
- P8_Gesamtende
Further tasks:
- checking primer with Geneious
- adapt primer to NEB-Phusion-polymerase conditions
2012-07-07
Primer-design for assembly-PCR, generating geneconstruct: scFv-TEV-TMD-EYFP
Investigators: Sascha
Aim: Design of primer for assembly-pcr of geneconstruct scFv-TEV-TMD-EYFP
Materials:
- Lablife Sequences of pcDNA5-FRT and pOG44
- Databases
- Oligocalc
- Wordpad
Method: forward sequencing using GATC-CMV-forward-primer
- P1_Signalp_N-term
- P2_Signalp_C-term
- P3_TMD-N-term
- P4_TMD-C-term/N-YFP
- P5_YFP-N-term/TMD-C-term
- P6_YFP-C-term
- P7_Gesamtanfang
- P8_Gesamtende
Further tasks:
- checking primer with Geneious
- adapt primer to NEB-Phusion-polymerase conditions
2012-07-10
Checking of Sequencing of GATC-results of pcDNA5-FRT and pOG44
Investigators: Sascha
Aim: Control of pcDNA5-FRT- and pOG44-sequences
Materials:
- Geneious
- Lablife Sequences of pcDNA5-FRT and pOG44
- GATC-viewer
- NCBI BLASTn
Results:
- sequences are correct
2012-07-12
ordering of primer for assembly-PCR, generating geneconstruct: scFv-TEV-TMD-EYFP
Investigators: Sascha, Maria
Aim: ordering of primer for assembly-pcr of geneconstruct scFv-TEV-TMD-EYFP
Materials:
- Lablife Sequences of pcDNA5-FRT and pOG44
- Databases
- Oligocalc
- Wordpad
- Geneious
- NEB-Calculator for high-fidelity Phusion-polymerase
Method: forward sequencing using GATC-CMV-forward-primer
- P1_Signalp_N-term
- P2_Signalp_C-term
- P3_TMD-N-term
- P4_TMD-C-term/N-YFP
- P5_YFP-N-term/TMD-C-term
- P6_YFP-C-term
- P7_Gesamtanfang
- P8_Gesamtende
Further tasks:
- extension-assembly-pcr
2012-07-15
Retransformation with scFv, transmembrane domain and YFP
Investigators: Maria
Aim: Retransformation with scFv anti-EGFR, transmembrane domain BBa_KI157010, YFP BBa_E0030
Date/Time: 15th July 2012, 2:30 – 4:30 pm
Materials: competent E. coli cells XL1 Blue, Biobricks BBa_K157010 and BBa_E0030, scFv anti-EGFR, 42°/37° C shaker, centrifuge
Method: see transformation protocol
Results: colonies on Amp plates with scFv-, transmembrane domain- and YFP- plasmid
Further tasks: picking clones and overnight culture (16th July)
2012-07-16
Topic: Overnight culture of scFv, YFP and transmembrane domain carrying cells
Investigators: Maria
Time: 2012-07-16; 6:30 - 7 pm
Materials:
- LB medium
- Amp. 25 mg/ ml stock solution
- Plasmids: scFv; BBa-KI57000 with transmembrane domain; pSB1AK3 with YFP
Method:
Inoculation of cell sample each in 5 ml LB medium with Amp
shaking over night at 37°C, 300 rpm, approx. 17 hours
Further tasks:
- Miniprep
2012-07-17
Topic: Mini Prep of YFP and transmembrane domain
Investigators: Stefan, Tarek, Kerstin
Time: 2012-07-17; 2 - 3.30 pm
Materials:
- overnight cultures from 2012-07-16
- GeneJET Plasmid Miniprep Kit (Thermo Scientific)
- Plasmids: BBa-KI57000 with transmembrane domain; pSB1AK3 with YFP
Method:
Miniprep according to Kit
Note:
overnight culture of retransformation of scFv did not grow --> Resistance was not known in scFv vector, screening was done with ampicillin
Further Tasks:
PCR to produce sp-scFv-tmd-yfp construct
2012-07-20
Topic: extension-assembly-pcr of scFv-TEV-TMD-EYFP geneconstruct
Investigators: Stefan, Sascha
Materials:
Materials:
scFv anti-EGFR, BBa-KI57000 with transmembrane domain; BBa_E0030 with EYFP
- P1_Signalp_N-term
- P2_Signalp_C-term
- P3_TMD-N-term
- P4_TMD-C-term/N-YFP
- P5_YFP-N-term/TMD-C-term
- P6_YFP-C-term
- P7_Gesamtanfang
- P8_Gesamtende
- Phusion -polymerase, dNTPs (10mM), Phusion HF-Buffer
Method:
- 50µl PCR mix of each plasmid= 10µl HF-Buffer; 1µl dNTPs(10mM); forward and reverse primer each 2,5µl (10µM); template DNA: 1µl of 200ng/µl scFv --> 200ng, 5µl of BBa_E0030 with EYFP (46,3ng/µl--> ca. 230ng),5µl of BBa-KI57000 with transmembrane domain (43,9ng/µl --> ca. 230ng); Phusion DNA polymerase 0,5µl; nclease-free water ad to 50µl
- PCR-program: initial Denat. 40´´ at 98°C; Denat 12´´ at 98°C; Annealing 15´´ at 55°C; Elongation 11´´ at 72°C; final-Elongation 10´at 72°C; 30 cycles
Further Tasks:
- gelextraction
- assembly-pcr
2012-07-26
Planning the antibody construct for genesynthesis
Investigators: Maria
Aim: construct with scFv 425bla, LoxP and TEV recognition site, E-YFP
Date/Time: 26th July 2012, 2 - 5 pm
Materials: Geneious
Results: antibody construct RCF25
Topic: extension-pcr of scFv-TEV-TMD-EYFP geneconstruct
Investigators: Sascha
Materials:
scFv anti-EGFR, BBa-KI57000 with transmembrane domain; BBa_E0030 with EYFP
- P1_Signalp_N-term
- P2_Signalp_C-term
- P3_TMD-N-term
- P4_TMD-C-term/N-YFP
- P5_YFP-N-term/TMD-C-term
- P6_YFP-C-term
- Phusion -polymerase, dNTPs (10mM), Phusion HF-Buffer
Method:
- 50µl PCR mix of each plasmid= 10µl HF-Buffer; 1µl dNTPs(10mM); forward and reverse primer each 2,5µl (10µM); template DNA: 1µl of 200ng/µl scFv--> 200ng, 5µl of BBa_E0030 with EYFP (46,3ng/µl--> ca. 230ng),5µl of BBa-KI57000 with transmembrane domain (43,9ng/µl --> ca. 230ng); Phusion DNA polymerase 0,5µl; nclease-free water ad to 50µl
- PCR-program: initial Denat. 40´´ at 98°C; Denat 12´´ at 98°C; Annealing 15´´ at 55°C; Elongation 11´´ at 72°C; final-Elongation 10´at 72°C; 30 cycles
Further Tasks:
- gelelectrophoresis
Topic: gelelectrophoresis of extended genes after extension-pcr
Investigators: Sascha
Aim: separation of extended genes scFv-TEV, TEV-TMD, EYFP in 1% agarosegel
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
- extended genes
Method:
- 1% agarosegel
- 70 min at 105V
Results:
Further Tasks:
- gelextraction
Topic: gelextraction of extended genes after extension-pcr
Investigators: Sascha
Aim: gelextraction of extended genes(scFv-TEV, TEV-TMD, EYFP) out of 1% agarosegel
Method:
- DNA extracted according to the manual
Results: concentration measuremnt using nanodrop
- concentrations of extended scFv:
- concentrations of extended TMD:
- concentrations of extended EYFP:
Further Tasks:
- assembly-pcr
Topic: assembly-pcr with extended genes to scFv-TEV-TMD-EYFP geneconstruct
Investigators: Sascha
Aim: assembly-pcr with extended genes to scFv-TEV-TMD-EYFP geneconstruct
Materials:
scFv anti-EGFR, BBa-KI57000 with transmembrane domain; BBa_E0030 with EYFP
- P7_Gesamtanfang (forward-primer)
- P8_Gesamtende (reverse-primer)
- Phusion -polymerase, dNTPs (10mM), Phusion HF-Buffer
Method:
- first assembly-pcr-mix--> 150:50: 10µl HF-Buffer; 1µl dNTPs(10mM); 5µl of extended TMD (30ng/µl --> 150ng), 5µl of extended EYFP (10ng/µl --> 50ng), 2µl of extended scFv (25ng/µl --> 50ng); nuclease-free water ad to 45µl; 0,5µl Phusion-polymerase
- second assembly-pcr-mix--> 90:30: 10µl HF-Buffer; 1µl dNTPs(10mM); 2,5µl of extended TMD (30ng/µl --> 90ng), 3µl of extended EYFP (10ng/µl --> 30ng), 1µl of extended scFv (25ng/µl --> 25ng); nuclease-free water ad to 45µl; 0,5µl Phusion-polymerase
- PCR-program to let the extended genes prime each other: initial Denat. 40´´ at 98°C; Denat 12´´ at 98°C; Annealing 15´´ at 55°C; Elongation 27´´ at 72°C; final-Elongation 10´at 72°C; 15 cycles
- after annealing of extended genes to each other 2,5µl of primer forw_P7_Gesamtanfang and primer rev_P8_Gesamtende were added
- PCR-prgram with end-primers: initial Denat. 40´´ at 98°C; Denat 12´´ at 98°C; Annealing 15´´ at 57°C; Elongation 27´´ at 72°C; final-Elongation 10´at 72°C; 27 cycles
- Gelelectrophoresis at 105V for 75`
Results:
- extraction of wrong 1100bp DNA-bond
Further Tasks:
- assembly-pcr
- extraction of correct DNA (1701bp)
2012-07-28
Topic: assembly-pcr with extended genes to scFv-TEV-TMD-EYFP geneconstruct
Investigators: Sascha, Tarek
Aim: assembly-pcr with extended genes to scFv-TEV-TMD-EYFP geneconstruct
Materials:
scFv anti-EGFR, BBa-KI57000 with transmembrane domain; BBa_E0030 with EYFP
- P7_Gesamtanfang (forward-primer)
- P8_Gesamtende (reverse-primer)
- Phusion -polymerase, dNTPs (10mM), Phusion HF-Buffer
Method:
- first assembly-pcr-mix--> 90:30: 10µl HF-Buffer; 1µl dNTPs(10mM); 2,5µl of extended TMD (30ng/µl --> 90ng), 3µl of extended EYFP (10ng/µl --> 30ng), 1µl of extended scFv (25ng/µl --> 25ng); nuclease-free water ad to 45µl; 0,5µl Phusion-polymerase
- second assembly-pcr-mix--> 140:20: 10µl HF-Buffer; 1µl dNTPs(10mM); 1µl of extended TMD (140ng/µl --> 140ng), 12,5µl of extended EYFP (1,6ng/µl --> 20ng), 4,8µl of extended scFv (4,2ng/µl --> ca. 20ng); nuclease-free water ad to 45µl; 0,5µl Phusion-polymerase
- PCR-program to let the extended genes prime each other: initial Denat. 40´´ at 98°C; Denat 12´´ at 98°C; Annealing 15´´ at 55°C; Elongation 27´´ at 72°C; final-Elongation 10´at 72°C; 15 cycles
- after annealing of extended genes to each other 2,5µl of primer forw_P7_Gesamtanfang and primer rev_P8_Gesamtende were added
- PCR-prgram with end-primers: initial Denat. 40´´ at 98°C; Denat 12´´ at 98°C; Annealing 15´´ at 57°C; Elongation 27´´ at 72°C; final-Elongation 10´at 72°C; 27 cycles
- Gelelectrophoresis at 105V for 85`
Results:
- extraction of correct DNA (1701bp)
Further Tasks:
- digestion of pcDNA5-FRT and geneconstruct scFv-TEV-TMD-EYFP
- ligation of plasmid and insert
2012-07-31
Planning and reviewing the antibody construct for genesynthesis
Investigators: Maria
Aim: construct with scFv 425bla, LoxP and TEV recognition site, mVenus
Date/Time: 27th July 2012, 3 - 5 pm
Materials: Geneious
Results: antibody construct RCF25
2012-07-31
Construct for genesynthesis with human scFv 425-72000
Investigators: Maria
Aim: construct with human scFv 425-72000, LoxP and TEV recognition site, mVenus
Date/Time: 27th July 2012, 3 - 5 pm
Materials: Geneious
Results: antibody construct RCF25
Virus
02.07.2012
Topic: PCR
Investigators: Xenia and Kathi
Aim:
- to test the primer
- amplification of the cap and VP-region, insertion of kozak-, Sortasemotive-, mycTag- and restiction sites Sequence
Materials:
- new primer combination (prr_VP2_pstI_Temp68 and prf_XbaI_kozak_So rtlN_myc_VP2 [c= 10 µM])
Method:
- polymerase chain reaction
Mastermix
| reagenz | volumen [µL] |
| HE buffer | 5 |
| dNTPs (NEB) | 1.25 |
| Primer (prr_VP2_pstI_Temp68) | 1.0 |
| Primer (prf_XbaI_kozak_So rtlN_myc_VP2) | 1.0 |
| DNA (Plasmid) | 1.0 |
| Phusion polymerase | 1.0 |
| water | 33.75 |
program
| step | temperature [°C] | duration [s] | cycles |
| denaturation | 95 | 120 | 1 |
| denaturation | 95 | 30 | 30 |
| annealing | 68 | 60 | 30 |
| elongation | 72 | 60 | 30 |
| final elongation | 72 | 60 | 1 |
| cooling | 4 | ∞ | 1 |
Results:
Further tasks:
- agarose gel electrophoresis
04.07.2012
Topic: gel electrophoresis
Investigator: Laura
Aim:
- measurement of DNA-concentration of pcr product and DARPin+cmv plasmid
- test pcr product and DRAPin+cmv plasmid
Materials:
- pcr product (02.07.) and DARPin+cmv Plasmid (Sven)
Method:
- agarose gel electrophoresis
- nanodrop
Results:
- DNA-concentrations:
- pcr-prduct: 266.1 ng/µL
- plasmid: 441.4 ng/µL
- gel electrophoresis
- pcr product present but concentration too high --> next time better: serial dilution of pcr product!
- pcr product present but concentration too high --> next time better: serial dilution of pcr product!
IMPORTANT!: Our primer is wrong! we have put the C-terminal sortase-motiv into our primer, but we just need the N-terminal sortase-tag (glycin-rests)
further tasks:
design new primer
04.07.2012
Topic: Primer design - Cloning: Change the C-terminal Sortase-Tag against the N-terminal
Investigators:Tobias and Xenia
Aim:
- Change the C-terminal sortase-tag against the N-terminal
Materials:
- prf_XbaI_kozak_SortaseMotivN_myc_VP2
Method: Geneious
Results:
- prf_Xba1 koz_GGGGG_VP2 (forward primer)
- prf_XbaI_koz_GGGGG_myc_VP2 (forward primer)
the sortase-tag was changed in both primers and prf_Xba1 koz_GGGGG_VP2 contain no myc-tag
Further tasks:
PCR
11.07.2012
Topic: PCR - Cloning: PCR using the new forward primers
Investigators: Kathi and Laura
Aim:
- PCR using the both new forward primer (prf_Xba1 koz_GGGGG_VP2 and prf_XbaI_koz_GGGGG_myc_VP2)
- gel electrophoresis
Materials:
- prf_Xba1 koz_GGGGG_VP2 (FP 1)--> myc-
- prf_XbaI_koz_GGGGG_myc_VP2(FP2)--> myc+
- prr_VP2_pstI_Temp68 (RP)
Method:
- PCR (PCR protocol: 02.07.2012)
- gel electropohoresis: : 2.1 and 0.5 µL pcr product of primer pair myc+ and myc- loaded on gel
Results:
- gel electrophoresis: pcr product at 2000 bp present, primer dimer obvious
Further tasks:
- ligation
12.07.2012
Topic: PCR
Investigators: Xenia and Laura
Aim:
- test the primer using different annealing temperatures to get the right sequence of 2000 bp
Materials:
primer combination:
- prr_VP2_pstI_Temp68 and prf_XbaI_koz_GGGGG_VP2 (FP1)-> myc-
- prr_VP2_pstI_Temp68 (RP) and prf_XbaI_koz_GGGGG_myc_VP2(FP2)--> myc+
Method:
- polymerase chain reaction
Mastermix
| reagenz | volumen [µL] |
| HF buffer | 5 |
| dNTPs (NEB) | 1.25 |
| Primer (prr_VP2_pstI_Temp68) | 1.0 |
| Primer (prf_XbaI_kozak_So rtlN_myc_VP2) | 1.0 |
| DNA (Plasmid) | 1.0 |
| Phusion Polymerase | 1.0 |
| water | 33.75 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| denaturation | 98 | 30 | 1 |
| denaturation | 98 | 10 | 30 |
| annealing | 64; 68; 70; 72 | 40 | 30 |
| elongation | 72 | 40 | 30 |
| final elongation | 72 | 300 | 1 |
| cooling | 4 | ∞ | 1 |
Results:
Further tasks:
- agarose gel electrophoresis
13.07.2012
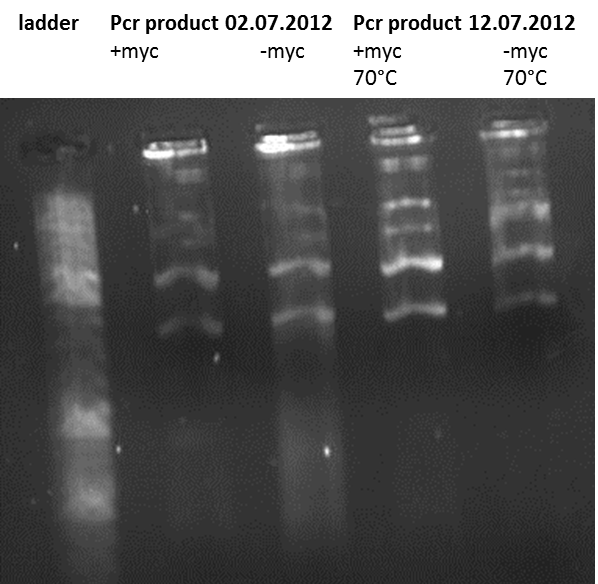
Topic: gel electrophoresis of the pcr product
Investigators:Xenia and Mario
Aim:gel electrophoresis of the pcr product (12.07.2012)
material and method:
- gel electrophoresis
- samples:
- 4 µL dest. Wasser + 1 µL pcr sample + 1 µL loading dye
- 4 µL dest. Wasser + 1 µL pcr sample + 1 µL loading dye
Results:
The fragment should have the size of 2000 bp, but the band is by 3000 bp
Further Tasks:
- test digestion
2012-17-07
Topic: test digestion
Investigators: Tobias
Materials:
- pcr products of 02-07 and 12-07-2012 (Ta= 70°C)
- restriction enzymes (FastDigest XbaI, PstI)
Method:
digestion: 10 µL DNA + 1 µL of each enzyme + 2 µL green-buffer + 17 µL water
Results:
- caused on the gel qualtity: repetition of the digestion
Further Tasks:
- Digestion
2012-18-07
Topic: preperative digestion an gel extraction of pcr fragments
Investigators: Tobias and Laura
Materials:
- pcr-products of 02-07-2012 and 12-07-2012 (Ta= 64 °C and 68 °C)
- restriction enzymes (FastDigest (FD) XbaI, PstI)
Method:
digestion: 10 µL DNA + 1 µL of each enzyme + 2 µL FD green-buffer + 17 µL water
Results:
2012-27-07
Topic: digestion of pcr fragments and plasmid
Investigators: Tobias
Materials:
- pcr-products of 12-07-2012
- plasmid (P10_pSB1C3_001_CMV_DARPin_ML_VP2/3_587KO_6xHis)
- restriction enzymes (FastDigest (FD) XbaI, PstI, SpeI)
Method:
digestion: 10 µL DNA + 1 µL of each enzyme + 2 µL FD green-buffer + 17 µL water
Results:
2012-01-08
Topic: Sequencing of pcr-pruducts
Investigator: Tobias
Materials:
- PCR-products of 12-07-2012 (+myc and-myc)
- Primer: prr_VP2_pstI_Temp68
Method:
GATC "
"