Team:Potsdam Bioware/Biobricks/Overview
From 2012.igem.org
(Difference between revisions)
(→Overview) |
(→Overview (click on a label to see the Bricks)) |
||
| (93 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
<div class="box_round gradient_grey"> | <div class="box_round gradient_grey"> | ||
==Biobricks== | ==Biobricks== | ||
| - | ===Overview=== | + | ===Overview (click on a label to see the Bricks)=== |
| - | + | <br> | |
| + | [[File:UP12biobricks.png|link=https://2012.igem.org/Team:Potsdam_Bioware/Biobricks/All_Biobricks|600px|left]] | ||
<html> | <html> | ||
| + | |||
| + | |||
| + | <div style="position:absolute; top:600px; left:730px;"> | ||
| + | |||
| + | |||
<script type="text/javascript" language="JavaScript"> | <script type="text/javascript" language="JavaScript"> | ||
<!-- Begin | <!-- Begin | ||
image1 = new Image; | image1 = new Image; | ||
| - | image1.src = "https://static.igem.org/mediawiki/2012/ | + | image1.src = "https://static.igem.org/mediawiki/2012/c/c6/Rfp2.png"; |
image2 = new Image; | image2 = new Image; | ||
| Line 22: | Line 28: | ||
</script> | </script> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | <a href="https://2012.igem.org/Team:Potsdam_Bioware/Biobricks/All_Biobricks#Potsdam_Standard_BioBricks" | ||
| + | onmouseover="button04.src=image2.src" | ||
| + | onmouseout="button04.src=image1.src"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/c/c6/Rfp2.png" name="button04" width="200" border="0"> | ||
| + | </a> | ||
| + | </div | ||
</html> | </html> | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | <div style="position:absolute; top:834px; left:575px;"> | ||
| + | <span style="color:#00C000; font-weight:bold; font-size: 20px;">[https://2012.igem.org/Team:Potsdam_Bioware/Biobricks/All_Biobricks#Selection_BioBricks Selection Parts]</span> | ||
| + | </div> | ||
| - | <div style="position:absolute; top: | + | <div style="position:absolute; top:493px; left:142px;"> |
| - | <span style="color:#00C000; font-weight:bold; font-size: 20px;">[ | + | <span style="color:#00C000; font-weight:bold; font-size: 20px;">[https://2012.igem.org/Team:Potsdam_Bioware/Biobricks/All_Biobricks#Antibody_BioBricks Antibody Parts]</span> |
</div> | </div> | ||
| - | <div style="position:absolute; top: | + | <div style="position:absolute; top:683px; left:168px;"> |
| - | <span style="color:#00C000; font-weight:bold; font-size: 20px;">[ | + | <span style="color:#00C000; font-weight:bold; font-size: 20px;">[https://2012.igem.org/Team:Potsdam_Bioware/Biobricks/All_Biobricks#Mutation_BioBricks Mutation Parts]</span> |
</div> | </div> | ||
| - | <div style="position:absolute; top: | + | <div style="position:absolute; top:575px; left:680px;"> |
| - | <span style="color:#00C000; font-weight:bold; font-size: 20px;">[ | + | <span style="color:#00C000; font-weight:bold; font-size: 20px;">[https://2012.igem.org/Team:Potsdam_Bioware/Biobricks/All_Biobricks#Potsdam_Standard_BioBricks RFC Potsdam Standard Parts]</span> |
</div> | </div> | ||
| - | + | ---- | |
===Favorite Biobricks=== | ===Favorite Biobricks=== | ||
| Line 50: | Line 62: | ||
<tr> | <tr> | ||
| - | <td>[http://partsregistry.org/Part: | + | <td>[http://partsregistry.org/Part:BBa_K929107 BBa_K929107]<br> |
| - | '''Anti-GFP, IgG1 Fc with TEV site, | + | '''Anti-GFP, IgG1 Fc with TEV site, TMD, mCherry, framed by LoxP'''<br> |
| - | + | ||
</td> | </td> | ||
| - | <td> [[Image: | + | <td> [[Image:UP12_BBa_K929107.png|right|400px]]This part was derived by de novo synthesis based on sequence information provided by GenBank and UniProt and by one already existing Biobrick part. The antibody construct consists of two major building blocks represented by the actual antibody unit (BBa_K929102) and the switchable membrane anchoring region (BBa_K929103). Both elements guarantee the eligibility and the easy handling of the construct and are optimized for expression in CHO cells. The antibody unit is represented by the human Ig kappa chain V-I region signal peptide (UniProt: P01601), the anti-GFP Nanobody (PDB: 3OGO) and the Fc region (UniProt: P01857). TEV protease recognition site, 2 LoxP sites, the B-cell receptor transmembrane domain (modified BBa_K157010) and the mCherry reporter display the switchable membrane anchoring region. The TEV recognition site on protein level and the LoxP sites on the genetically level allow the shift from surface presentation to secretion of the antibody unit. For detailed information see the [http://partsregistry.org/Part:BBa_K929107 partsregistry], for experimental experience also see [https://2012.igem.org/Team:Potsdam_Bioware/Biobricks/All_Biobricks All Biobricks]. |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 61: | Line 72: | ||
<td>[http://partsregistry.org/Part:BBa_K929003 BBa_K929003]<br> | <td>[http://partsregistry.org/Part:BBa_K929003 BBa_K929003]<br> | ||
'''modified AID with CMV, hGH-polyA and eGFP'''<br> | '''modified AID with CMV, hGH-polyA and eGFP'''<br> | ||
| - | |||
</td> | </td> | ||
<td> [[Image:UP12_BBa_K929003_smaloverview.png|right|400px]] This part is an improved version of wildtype AID, the enzyme that randomly mutates predominantly in the immunoglobulin genes. It is designed for strong expression of the fusion protein modified AID+eGFP. Modified AID has an additional Nuclear Localization Sequence (NLS)and the naturally occurring Nuclear Export Sequence (NES)is deleted. Due to the fact that AID mutates the actively transcribed single stranded DNA, it is supposed that the direction of the enzyme to the inside of the nucleus would improve the mutation rate.The fusion with the green fluorescent reporter eGFP allows 1.)to check the transfection success 2.)select transfected cells via FACS and 3.)to check the cellular localization of the fusion protein. For detailed information see the [http://partsregistry.org/Part:BBa_K929003 partsregistry], for experimental experience also see [https://2012.igem.org/Team:Potsdam_Bioware/Biobricks/All_Biobricks All Biobricks]. | <td> [[Image:UP12_BBa_K929003_smaloverview.png|right|400px]] This part is an improved version of wildtype AID, the enzyme that randomly mutates predominantly in the immunoglobulin genes. It is designed for strong expression of the fusion protein modified AID+eGFP. Modified AID has an additional Nuclear Localization Sequence (NLS)and the naturally occurring Nuclear Export Sequence (NES)is deleted. Due to the fact that AID mutates the actively transcribed single stranded DNA, it is supposed that the direction of the enzyme to the inside of the nucleus would improve the mutation rate.The fusion with the green fluorescent reporter eGFP allows 1.)to check the transfection success 2.)select transfected cells via FACS and 3.)to check the cellular localization of the fusion protein. For detailed information see the [http://partsregistry.org/Part:BBa_K929003 partsregistry], for experimental experience also see [https://2012.igem.org/Team:Potsdam_Bioware/Biobricks/All_Biobricks All Biobricks]. | ||
| Line 72: | Line 82: | ||
<td>[http://partsregistry.org/Part:BBa_K929301 BBa_K929301]<br> | <td>[http://partsregistry.org/Part:BBa_K929301 BBa_K929301]<br> | ||
'''Potsdam Standard Backbone'''<br> | '''Potsdam Standard Backbone'''<br> | ||
| - | |||
</td> | </td> | ||
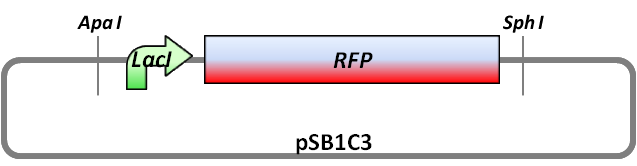
| - | <td> [[Image:UP12_BBa_K929301_smaloverview.png|right|400px]] This part is | + | <td> [[Image:UP12_BBa_K929301_smaloverview.png|right|400px]] This part is the new backbone of the Potsdam Standard. It contains two new restriction sites, Apa I and Sph I, and a RFP expression cassette which can be used as a ligation control. For the cloning process, the backbone has to be digested with Apa I and Sph I. The insert is amplified with primers which contains thiophosphates at the 5' end. After knocking out the thiophosphates with iodine/ethanol solution, the insert can be ligated into the digested backbone. The ligation success can be controlled by the red fluorescent protein. If the ligation faild, you would see red fluorescent colonies. For detailed information see the [http://partsregistry.org/Part:BBa_K929301 partsregistry], for experimental experience also see [https://2012.igem.org/Team:Potsdam_Bioware/Biobricks/All_Biobricks All Biobricks]. |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 83: | Line 92: | ||
| - | + | </div> | |
| - | + | ||
Latest revision as of 17:08, 26 October 2012
Biobricks
Overview (click on a label to see the Bricks)
Favorite Biobricks
| [http://partsregistry.org/Part:BBa_K929107 BBa_K929107] Anti-GFP, IgG1 Fc with TEV site, TMD, mCherry, framed by LoxP |
This part was derived by de novo synthesis based on sequence information provided by GenBank and UniProt and by one already existing Biobrick part. The antibody construct consists of two major building blocks represented by the actual antibody unit (BBa_K929102) and the switchable membrane anchoring region (BBa_K929103). Both elements guarantee the eligibility and the easy handling of the construct and are optimized for expression in CHO cells. The antibody unit is represented by the human Ig kappa chain V-I region signal peptide (UniProt: P01601), the anti-GFP Nanobody (PDB: 3OGO) and the Fc region (UniProt: P01857). TEV protease recognition site, 2 LoxP sites, the B-cell receptor transmembrane domain (modified BBa_K157010) and the mCherry reporter display the switchable membrane anchoring region. The TEV recognition site on protein level and the LoxP sites on the genetically level allow the shift from surface presentation to secretion of the antibody unit. For detailed information see the [http://partsregistry.org/Part:BBa_K929107 partsregistry], for experimental experience also see All Biobricks. |
| [http://partsregistry.org/Part:BBa_K929003 BBa_K929003] modified AID with CMV, hGH-polyA and eGFP |
This part is an improved version of wildtype AID, the enzyme that randomly mutates predominantly in the immunoglobulin genes. It is designed for strong expression of the fusion protein modified AID+eGFP. Modified AID has an additional Nuclear Localization Sequence (NLS)and the naturally occurring Nuclear Export Sequence (NES)is deleted. Due to the fact that AID mutates the actively transcribed single stranded DNA, it is supposed that the direction of the enzyme to the inside of the nucleus would improve the mutation rate.The fusion with the green fluorescent reporter eGFP allows 1.)to check the transfection success 2.)select transfected cells via FACS and 3.)to check the cellular localization of the fusion protein. For detailed information see the [http://partsregistry.org/Part:BBa_K929003 partsregistry], for experimental experience also see All Biobricks. |
| [http://partsregistry.org/Part:BBa_K929301 BBa_K929301] Potsdam Standard Backbone |
This part is the new backbone of the Potsdam Standard. It contains two new restriction sites, Apa I and Sph I, and a RFP expression cassette which can be used as a ligation control. For the cloning process, the backbone has to be digested with Apa I and Sph I. The insert is amplified with primers which contains thiophosphates at the 5' end. After knocking out the thiophosphates with iodine/ethanol solution, the insert can be ligated into the digested backbone. The ligation success can be controlled by the red fluorescent protein. If the ligation faild, you would see red fluorescent colonies. For detailed information see the [http://partsregistry.org/Part:BBa_K929301 partsregistry], for experimental experience also see All Biobricks. |
 "
"