Team:Paris Bettencourt/SID
From 2012.igem.org
Zmarinkovic (Talk | contribs) (→Overview) |
Zmarinkovic (Talk | contribs) (→Overview) |
||
| Line 7: | Line 7: | ||
Provide an overview of this sub-project in a couple of phrases | Provide an overview of this sub-project in a couple of phrases | ||
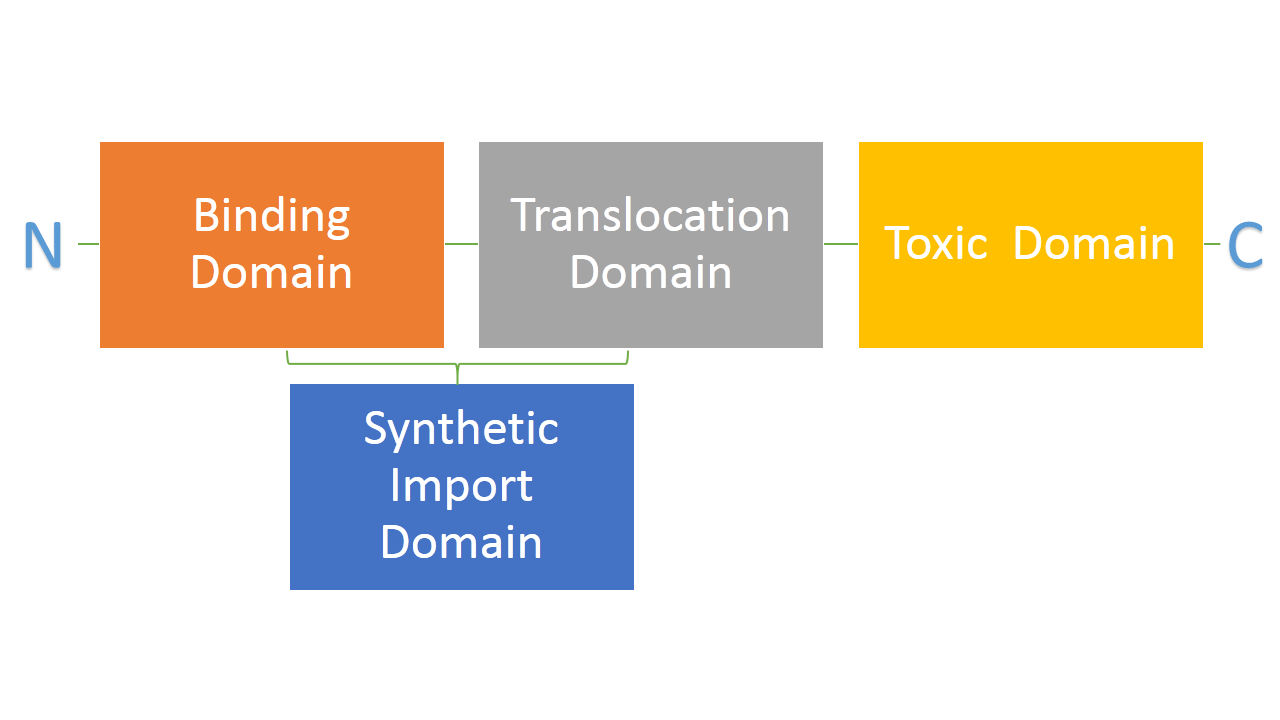
| + | Bacteria developed mechanisms to kill other bacteria, thus reducing competition between them in the environment. Some strains of E. coli produce lethal proteins called colicins which kill other bacteria, including E. coli. Colicins are built out of three main domains which are also a point of difference among many types of colicins. First domain is responsible for binding to a receptor on a bacterial membrane, second one is responsible for translocating the protein from outside to inside of a bacteria and the third one is responsible for killing bacteria. We are interested in two types of colicins, colicin E2 and colicin D. Both of them use different binding, translocation and killing mechanisms. Colicin E2 is a DNAse and colicin D is an RNAse. | ||
| - | + | We hypothesize that it is possible to use binding and translocation part of colicin as a "synthetic import domain" onto which we can fuse other proteins in order to import them into bacteria. Main aim of this part of the project is to fuse the RNAse domain from colicin D to the "synthetic import domain" of colicin E2. By doing this we will obtain a toxin that targets the same receptor and translocation mechanism but kills bacteria on the level of protein synthesis by cleaving tRNA. [link to suicide group]. | |
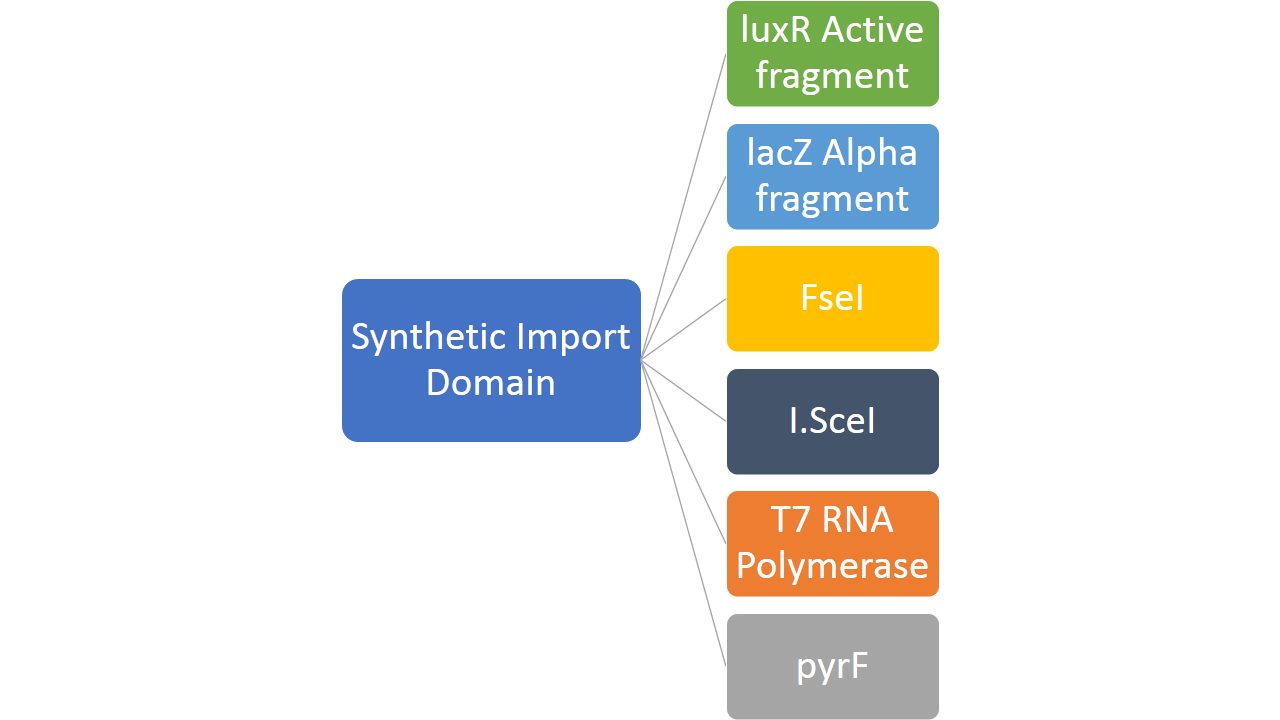
| - | + | In addition to this, we fused restriction enzymes FseI and I-SceI both to the colicin E2 and colicin D "synthetic import domain". These two enzymes are used in different parts of our project [link to restriction group]. We also phused a couple of other proteins to easily test the plausibility of our "synthetic import domain" system. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | In addition to this, we | + | |
==Objectives== | ==Objectives== | ||
Revision as of 16:02, 24 September 2012
Contents |
Overview
Provide an overview of this sub-project in a couple of phrases
Bacteria developed mechanisms to kill other bacteria, thus reducing competition between them in the environment. Some strains of E. coli produce lethal proteins called colicins which kill other bacteria, including E. coli. Colicins are built out of three main domains which are also a point of difference among many types of colicins. First domain is responsible for binding to a receptor on a bacterial membrane, second one is responsible for translocating the protein from outside to inside of a bacteria and the third one is responsible for killing bacteria. We are interested in two types of colicins, colicin E2 and colicin D. Both of them use different binding, translocation and killing mechanisms. Colicin E2 is a DNAse and colicin D is an RNAse.
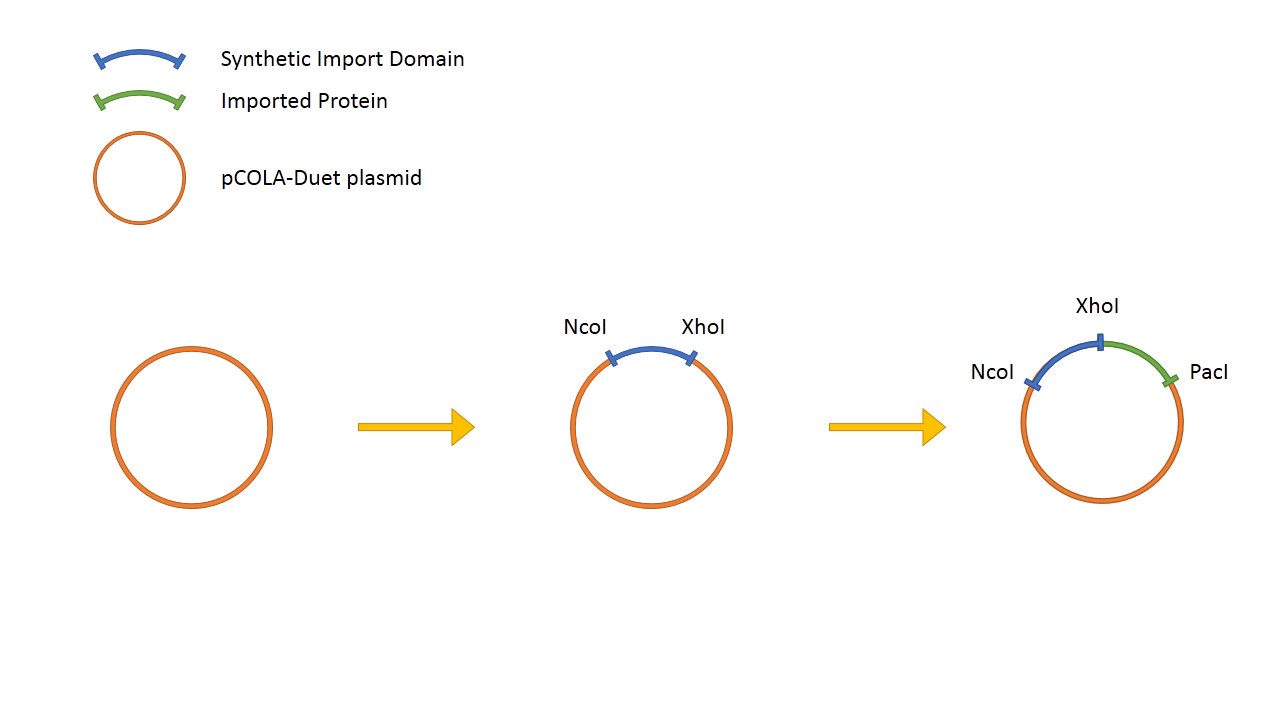
We hypothesize that it is possible to use binding and translocation part of colicin as a "synthetic import domain" onto which we can fuse other proteins in order to import them into bacteria. Main aim of this part of the project is to fuse the RNAse domain from colicin D to the "synthetic import domain" of colicin E2. By doing this we will obtain a toxin that targets the same receptor and translocation mechanism but kills bacteria on the level of protein synthesis by cleaving tRNA. [link to suicide group].
In addition to this, we fused restriction enzymes FseI and I-SceI both to the colicin E2 and colicin D "synthetic import domain". These two enzymes are used in different parts of our project [link to restriction group]. We also phused a couple of other proteins to easily test the plausibility of our "synthetic import domain" system.
Objectives
Write here the objectives of your project
Design
Present the design of your system, both in a written form, and a schematic one.
Experiments and results
Characterisation of X
Experimental setup
Describe the experiment
Results
Present your results
Testing of the system
Experimental setup
Describe the experiment
Results
Present your results
 "
"


 Overview
Overview Delay system
Delay system Semantic containment
Semantic containment Restriction enzyme system
Restriction enzyme system MAGE
MAGE Encapsulation
Encapsulation Synthetic import domain
Synthetic import domain Safety Questions
Safety Questions Safety Assessment
Safety Assessment