Team:OUC-China/Project/Sensor/GoalandDesign
From 2012.igem.org

Goal
What we desire to achieve is to characterize and modify the naturally existing sensors in prokaryotes in order to alarm the occurrence of red tide precisely and accurately. At the same time, try to find a way to restraint it. See more.
Design

Fig. The flowchart of our design is on the right side. To characterize the original promoter is the essential part for understanding the original promoter. After finishing this process, we continue to select engineering methods as well as seek for suggestion from our modeler. Experimental test goes along with the design.
promoter testing platform
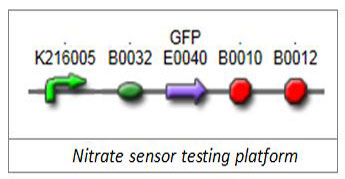
1.1 Nitrate-sensitive promoterWe firstly selected well-characterized promoter BBa_K216005 (Edinburgh 2009) as our nitrate-sensitive promoter. To verify its responding characterization, we chose green fluorescent protein (GFP) as our reporter and put its generator E0240 downstream of BBa_K216005.

1.2 Phosphate-sensitive promoter

phoBR promoter
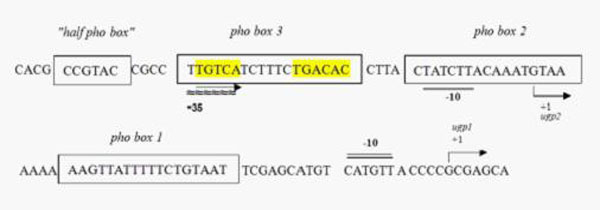
Ugp promoter
Since BBa_K116401 (NYMU-Taipei 2008) has not been submitted physically, we obtained two pho-box-contained promoters by PCR from strains E.coli-K12 MG1655. One of them is the phoBR promoter which contains one single pho-box. The other is the ugp promoter which contains several pho-boxes as well as CRP binding site associated with carbon metabolism. The latter one facilitates more variable phenotypes when introduced to multiple mutants by error-prone PCR. Besides, C:P is another important factors associated with red tide and we speculate that it may show great potential for further modifications. Similarly we have constructed test platforms for both of them by connecting E0240 downstream of each promoter.

Characterization
Phosphate-sensitive promoter characterization is somewhat difficult. Only when phosphate concentration is below 4uM could there be response. However, it is conflicting that phosphate is essential for bacteria to survive. Moreover, many complex medium, like LB medium, could not fit this requirement. So strict control of medium components is in need. Finally, we singled out the MOPS minimal medium designed by Frederick C.Neidhardt in 1974 [4] which is a kind of synthetic medium with MOPS as buffer agent rather than KHPO4/KH2PO4.
We developed a protocol for low-phosphate induction experiment designed specifically for phosphate-sensitive promoter characterization.See more.
We appled the real-time plate reader as our testing tool to detect whether the promoter works or not, ultra-sensitively respond or graded respond.
To learn more information please click here.
Promoter engineering methods
3.1 Overview
According to the papers and our characterization results, we realized that modifications of both phosphate- and nitrate-sensitive promoter are necessary when integrated into our whole project. The modification of former one is especially vital because the order of magnitude between susceptibility thresholds of BBa_K216005 and desirable one is nearly 100 times. To solve this question, we have collaborated with our modelers to extract the key factors from two corresponding TCSs. In addition, we conceived several potential solutions to further improve our responding sensitivity if time permits.
3.2 Affinity of response regulator(RR) binding site

To fine-tune RR binding site is one of the promoter engineering process. The detailed Flow chart can be seen on the right site. Error-Prone PCR could provide us with a large-scale promoter library which could be used as the ‘seed bank’. We could screen several ones which meet our needs.
We have tried to construct the promoter libraries of PphoBR, Pugp and K216005 by error-prone PCR. Unfortunately, we did not get the ideal promoter library, further experiment is still in progress.
3.3 the number of histone kinase (HK) molecules


Lu Zhou’s experiment has proved that HK concentration could influence the proportion and sensitivity of stimulated cells. Curve colors represent different strengths of induced HK expression[5]
It is interesting to find that TCS is highly homologous system. Firstly, both HK and RR coding genes share some highly similar motif. Secondly, HK and RR coding genes in each TCS are all transcribed in the same polycistron. Moreover, it has been proved that HK translation rates are significantly lower than RR which results in all-or-none behavior. This kind of behavior shows high stochastic noise which seriously interferes function of the system. So we construct an amplifier to raise the number of HK molecules to ensure the robustness and sensitivity.
To verify the feasibility of this method, we firstly constructed an amplifier composed of constitutive promoter J23106 and HK phoR. If our amplifier could increase the fluorescence value, we could confirm that it works as expected. The experiment is still in progress due to time limit.
3.4Construct of chimeric chemoreceptor & RBS selection for appropriate expression rate
Since our experiment hasn’t got to this step, we will discuss it in our future work column.
Modeling
Since our modelers have some problem with the construction of complex topological network as well as limited time, we took modeling examples by Kierzek, A. M[5] for better design. Due to the notable phenomenon of relatively high stochastic noise in TCS, we chose the Gillespie algorithm to simulate the whole reaction process involved in TCS. The table with empirical constants and propensity functions of TCS is shown here. and we try to change the constants of translational and transcriptional process for better understanding of the mechanisms.

Fig. This is the Gillepse’s algorithm simulation of the variation of reporter genes with changing time.The x axis represents the time range and the y axis represents the molecules of reporter. (A)was run under 0.8 transcription rate of HK which is similar to the situation where J23106 is upstream of HK generator. (B)was run under 0.3 transcription rate of HK. Both of them are assumed to accept the same intensity of environmental signals by the same ratio between phosphorylated HK/non-phosphorylated one. The results show that higher HK transcription rate, which suggests higher HK molecules in the cell, exerts significant influence on the responding time and stochastic noise of the system. (A) with much more HK molecules show shorter inducing time , higher reporter molecules and more induced cells when compared with (B). It is a strong evidence for the feasibility of constructing a HK amplifier to reduce stochastic noise and enhance the response speed.
Designing schemes
Although there are still a lot of works to do with phosphate- and nitrate-sensitive sensor, we have established an essential design to link up with ratio sensor. After discussing with my team member, I proposed that the phosphate sensor should be biobricked with an inverter to modulate the responding curve coupled with nitrate sensor. The reason is that phosphate-sensitive promoter is low-phosphate activated while transcription rate of nitrate-sensitive promoter is in proportion to the extracellular concentration of nitrate. Since the two signal inputs of ratio sensor should be in synchronized, reversal of the behavior of phosphate-sensitive promoter is needed. Here is our preliminary design:

Construction schemes:
Phosphate sensor device:

phosphate accumulated device
Researchers have demonstrated Strain MV1184 containing ppk and ackA showed the highest rates of Pi removal from the medium. This recombinant strain removed approximately 90% of the Pi from the medium within 4 h. The data of this result is very satisfactory. Although another recombination strain contain ppk and pst cluster has better effect, the data is not available, so we choose to creat the recombination strain containing ppk and ackA that we standardize by ourselves.
To get standardized parts, an entire ackA gene fragment was amplified by PCR with the primers designed by myself from the mutation of E.coli K12 strain ,DH5αand inserted into the PSB1C3 vector. Ppk contains an EcoRI site need to be mutant. So we get it from the E. coli DH5 Chromosome and inserted into pGEM-T-vector, then cut it in XbalI and PstI and inserted the fragment into PSB1C3 cutting by XbalI and PstI sites first.
 "
"
 Home
Home
 HumanPractice index
HumanPractice index
 JudgingForm
JudgingForm
 Contact Us
Contact Us
 Project Overview
Project Overview
 Sensor
Sensor
 Decision-making Device
Decision-making Device
 Gas vesicle
Gas vesicle
 ODEModel
ODEModel
 Parameter Sensitivity Analysis
Parameter Sensitivity Analysis
 Parameter Sweep
Parameter Sweep
 Noise Analysis
Noise Analysis
 HumanPractice Overview
HumanPractice Overview
 Meeting and Academic Communication
Meeting and Academic Communication
 Camps, Class and Lectures
Camps, Class and Lectures
 Special HP
Special HP
 Team Members
Team Members
 Instructors
Instructors
 Acknowledgement&Cooperation
Acknowledgement&Cooperation
 Lab
Lab
 Parts
Parts
 Safety
Safety
 Labnote
Labnote
 Modeling Note
Modeling Note
 Protocols
Protocols

