Team:NTU-Taida/Project/Stability
From 2012.igem.org
| Line 12: | Line 12: | ||
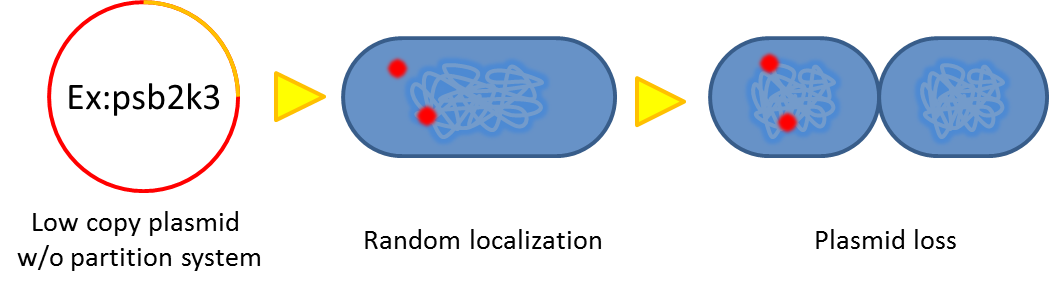
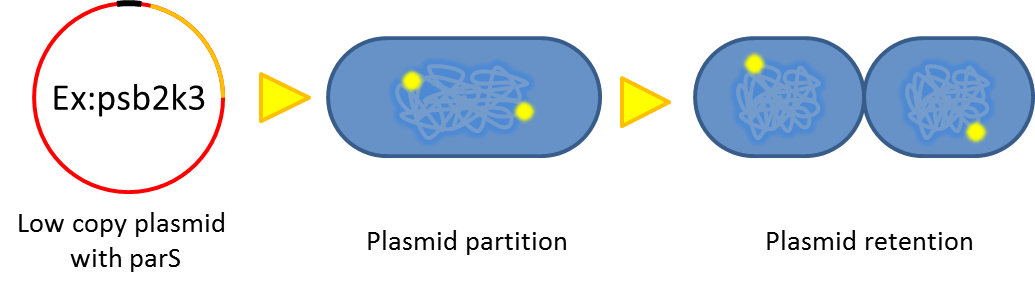
# Segregational instability <p style="text-indent: 2em;"> Plasmids are randomly (possibly uneven) distributed inside a bacterium. After cell division, one of the progenies may lose plasmids.</p>[[File:NTU-Taida-Par1.png|500px|center]] | # Segregational instability <p style="text-indent: 2em;"> Plasmids are randomly (possibly uneven) distributed inside a bacterium. After cell division, one of the progenies may lose plasmids.</p>[[File:NTU-Taida-Par1.png|500px|center]] | ||
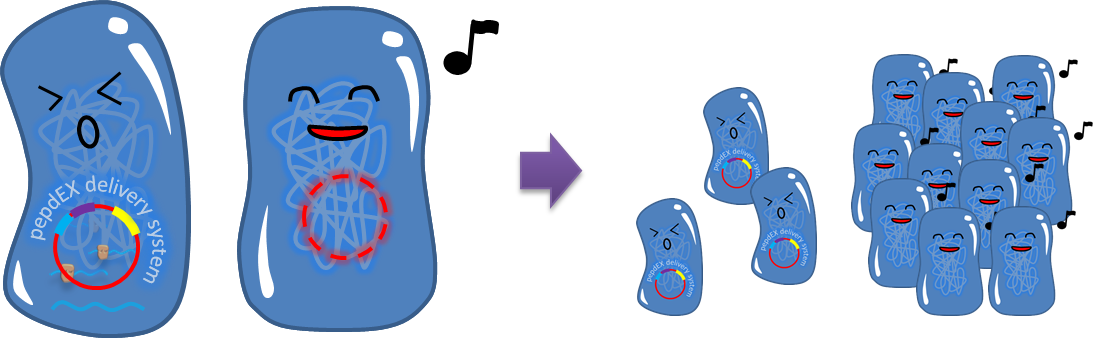
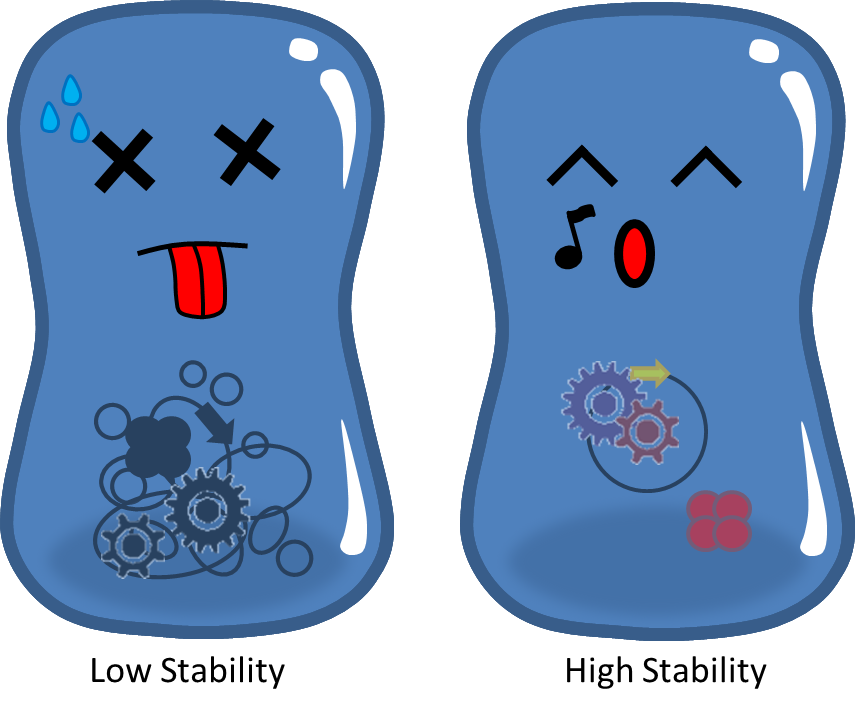
# Burden Effect <p style="text-indent: 2em;">Cells bearing our PepdEx system have to spend extra energy/resource, so will not grow as good as plasmid-free cells. This metabolic burden will cause our coli to gradually lose pieces of circuits, and the growth rate difference will eventually eliminate plasmid-bearing bacteria in the population.</p>[[File:NTU-Taida-Burden3.png|500px|center]] | # Burden Effect <p style="text-indent: 2em;">Cells bearing our PepdEx system have to spend extra energy/resource, so will not grow as good as plasmid-free cells. This metabolic burden will cause our coli to gradually lose pieces of circuits, and the growth rate difference will eventually eliminate plasmid-bearing bacteria in the population.</p>[[File:NTU-Taida-Burden3.png|500px|center]] | ||
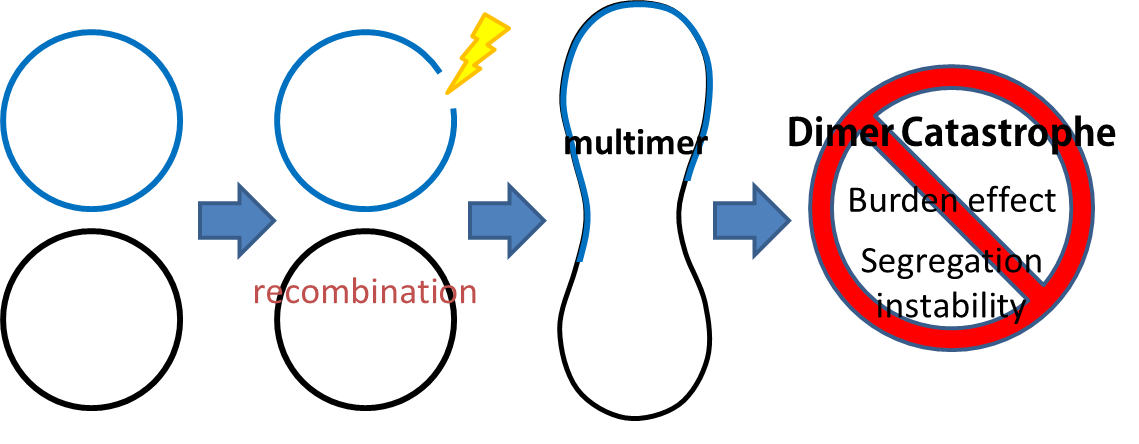
| - | # Dimer Catastrophe<p style="text-indent: 2em;">Homologues recombination may cause plasmid multimers which will increases segregational instability and burden[[# | + | # Dimer Catastrophe<p style="text-indent: 2em;">Homologues recombination may cause plasmid multimers which will increases segregational instability and burden[[#Ref|[1] ]]. This is the reason why most lab coli are recA1 mutants. But since our coli must be able to colonize in the gut, it should be recA+ wild type strain. That's the problem!</p>[[File:NTU-Taida-Mrs-1.png|500px|center]] |
===Stabilization Modules=== | ===Stabilization Modules=== | ||
| Line 20: | Line 20: | ||
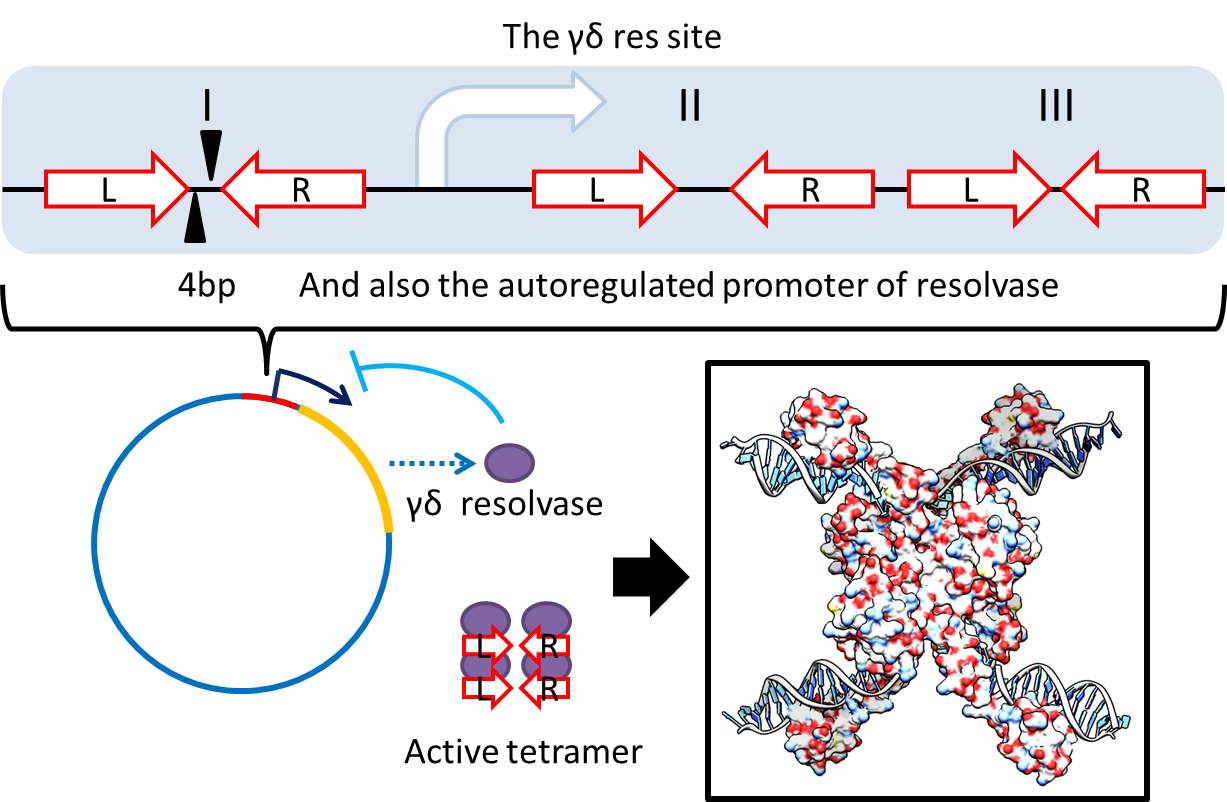
[[FIle:NTU-Taida-Mrs-2.png|600px|center|structure of MRS]] | [[FIle:NTU-Taida-Mrs-2.png|600px|center|structure of MRS]] | ||
| - | :<p style="text-indent: 2em;">To deal with multimerization, we cloned an cassette (BBa_K817018) which encodes an autoregulated resolves (serine type recombinase) from E.coli F plasmid transposon tn1000 (tn3 family)[[# | + | :<p style="text-indent: 2em;">To deal with multimerization, we cloned an cassette (BBa_K817018) which encodes an autoregulated resolves (serine type recombinase) from E.coli F plasmid transposon tn1000 (tn3 family)[[#Ref|[2] ]]. Its promoter region consists of 3 sub-sites(res site) and can process recombination[[#Ref|[3] ]]. This cassette can resolve multimer that formed during replicative transposition[[#Ref|[4] ]]. It can also resolve plasmid multimer to increase plasmid stability by avoiding dimer catastrophe. Multimer resolution system (MRS) provides analogous functions to E.coli chromosome XerCD/dif but acts independent of cell cycle, DNA localization and may have higher efficiency on plasmids compared with slow XerCD system[[#Ref|[5] ]].</p> |
[[FIle:NTU-Taida-Mrs-4.png|200px|center]] | [[FIle:NTU-Taida-Mrs-4.png|200px|center]] | ||
| Line 28: | Line 28: | ||
====2. Partition System:from '''<em>Pseudomonas putida''' KT2440</em>==== | ====2. Partition System:from '''<em>Pseudomonas putida''' KT2440</em>==== | ||
| - | :<p style="text-indent: 2em;">Bacterium distributes chromosomes/plasmids evenly to its progenies by a dynamic system that includes SMC like proteins, type Ia partition system and so on[[# | + | :<p style="text-indent: 2em;">Bacterium distributes chromosomes/plasmids evenly to its progenies by a dynamic system that includes SMC like proteins, type Ia partition system and so on[[#Ref|[6] ]]. Type Ia partition system segregates chromosomes/plasmids in a process akin to Eukaryotic mitosis. It can be found on most eubacteria. However, E. coli is not the case.</p> |
[[FIle:NTU-Taida-Par2.png|700px|center]] | [[FIle:NTU-Taida-Par2.png|700px|center]] | ||
| - | :<p style="text-indent: 2em;">Previous study have shown that when providing parAB of P.putida in trans and carrying parS site in cis, segregation of low copy plasmids (mini F) can be stabilized in E. coli.[[# | + | :<p style="text-indent: 2em;">Previous study have shown that when providing parAB of P.putida in trans and carrying parS site in cis, segregation of low copy plasmids (mini F) can be stabilized in E. coli.[[#Ref|[7] ]]]].The BioBrick vector pSB2K3 is a mini F plasmid, thus we expect we can stabilize its segregation this way. Our team have made a modified version of pSB2K3 with parS site (BBa_K817017), and plan to use it to harbor our PepdEx system (a low copy number plasmid allows us to fine tune our systems controls e.g. with smaller number of operator sites). We have also cloned the parAB operon (BBa_K817019), and provided the registry with the short 16bp parS site (BBa_K817016) for future applications.</p> |
[[FIle:NTU-Taida-Par3.png|500px|center]] | [[FIle:NTU-Taida-Par3.png|500px|center]] | ||
| Line 47: | Line 47: | ||
|} | |} | ||
| - | :<p style="text-indent: 2em;">Type I toxin-antitoxin srnBC is an ideal executor which belongs to hok/sok homologues. It expresses stable toxin encoding RNA and short-lived antitoxin RNA that can neutralize toxin RNA by RNA interaction and RNase III cleavage[[# | + | :<p style="text-indent: 2em;">Type I toxin-antitoxin srnBC is an ideal executor which belongs to hok/sok homologues. It expresses stable toxin encoding RNA and short-lived antitoxin RNA that can neutralize toxin RNA by RNA interaction and RNase III cleavage[[#Ref|[9] ]]. It acts as post segregation killing system – a bacterial cell will get killed if it loses the DNA (genomic islands, plasmids, mobile gene elements) that contains the module[[#Ref|[10] ]]. We plan to use it to reduce plasmid-lost cells in our coli population, in the hope to make applications without antibiotic selection more feasible. We have cloned the cassette with BioBrick standard (BBa_K817015). Following pictures show the mechanism (based on hok/sok).</p> |
| - | [[FIle:NTU-Taida-SrnBC-Mechanism1.png|600px|thumb|center|<p style="text-indent: 2em;">TA loci express two kinds of RNA, namely srnB and srnC, respectively. Initially, srnB is inactive and stable (half-life in the order of 30 minutes [[# | + | [[FIle:NTU-Taida-SrnBC-Mechanism1.png|600px|thumb|center|<p style="text-indent: 2em;">TA loci express two kinds of RNA, namely srnB and srnC, respectively. Initially, srnB is inactive and stable (half-life in the order of 30 minutes [[#Ref|[11] ]]) and will gradually be processed to active form producing hydrophobic toxic protein if srnC RNA does not exist; srnC is relative unstable and is degraded quickly, and it overlaps and compliments to 5’sequence of srnB.</p>]] |
| - | [[FIle:NTU-Taida-SrnBC-Mechanism2.png|600px|thumb|center|<p style="text-indent: 2em;">At normal condition (plasmid retained), srnC can pair with processed srnB and forms double stranded RNA that is susceptible to RNase III cleavage and thus prevents toxin production. If the plasmid loses, srnC will quickly disappear without replenishing and srnB will show its power[[# | + | [[FIle:NTU-Taida-SrnBC-Mechanism2.png|600px|thumb|center|<p style="text-indent: 2em;">At normal condition (plasmid retained), srnC can pair with processed srnB and forms double stranded RNA that is susceptible to RNase III cleavage and thus prevents toxin production. If the plasmid loses, srnC will quickly disappear without replenishing and srnB will show its power[[#Ref|[12] ]].</p>]] |
====4. Reduce Burden Effect==== | ====4. Reduce Burden Effect==== | ||
| Line 58: | Line 58: | ||
<p style="text-indent: 2em;">High level gene expression and gene dosage leads to burden effect. Therefore, if two systems are encoded in different plasmids with the same overall performance, low copy plasmids are preferred over high copy ones. Again if with the same overall outcome, stabilizing mRNA is preferred over overexpressing the gene (unless for regulatory purposes). Place yourselves in E. coli's position! Reduce its metabolic burden as much as possible then it will work for you! </p> | <p style="text-indent: 2em;">High level gene expression and gene dosage leads to burden effect. Therefore, if two systems are encoded in different plasmids with the same overall performance, low copy plasmids are preferred over high copy ones. Again if with the same overall outcome, stabilizing mRNA is preferred over overexpressing the gene (unless for regulatory purposes). Place yourselves in E. coli's position! Reduce its metabolic burden as much as possible then it will work for you! </p> | ||
| - | |||
[[FIle:NTU-Taida-Burden2.png|330px|center]] | [[FIle:NTU-Taida-Burden2.png|330px|center]] | ||
| - | == | + | == Safety of Delivery System == |
| - | < | + | <p style="text-indent: 2em;">Many turn off strategies have been developed, most of them are inducible suicide systems that can be activated at certain conditions. In our project, we plan to use temperature as the activating signal (or alternatively, small molecules). When the course of treatment ends, leaving human body (temperature drop from 37C to room temperature) or administration of small amount of drugs (e.g. tetracycline) will cause suicide gene activation, thereby avoiding recombinant strain/gene pollution. And splitting suicide system to provide repression in trans can prevent plasmid transferring to wild type strains. Many off-the-rack parts can be used.</p> |
| - | + | ||
| - | + | [[File:NTU-Taida-Suicide-system.png|300px|thumb|left|alt=new idea about safety|general scheme of suicide system ]] | |
| - | < | + | |
| - | + | <p style="text-indent: 2em;">However, these designs cannot completely eliminate the risk of horizontal gene transfer (HGT), where recombinant genes can move to other organisms independent of suicide system. Therefore, in addition to suicide system, we propose a new concept to deal with HGT risks by RNA interaction[[#Ref|[1] ]]. Although bacteria lack RNAi pathway, expressing well designed antisense RNAs have been shown to have inhibitory effect on target RNAs through competitive inhibition.A recent study has demonstrated that expressing peptide nucleic acid (PNA) antisense to antitoxin RNA 5' sequence kills bacteria cells [[#Ref|[2] ]]. We reason that adding rationally designed antisense RNAs on the untranslated region of transcripts may interfere with target RNA’s function or translation, and we can employ this property to prevent HGT. For instance, HGT is more likely to occur between related species, such as lab E. coli & O157. Laboratory E. coli have inactivated all its hok/sok toxin-antitoxin system by mutation, but wild bacteria (especially pathogenic bacteria) usually have more active TA locus on its chromosome like E. coli O157. We plan to put a stem loop from hok mRNA, which can pair with sok RNA, 5’sequence on UTR of antibiotic resistance genes used in our PepdEx system. IF wild bacteria steal our antibiotic resistance genes and express it, its antitoxin will be competitive inhibited, and its toxin will get expressed to kill the thief. This prevents HGT between lab & wild coli. This concept can have wide extension. In addition to targeting antitoxin (functional RNA) of type I TA, it is also possible to design antisense sequences that target RBS to down-regulate targeted protein. Targeting antitoxin of type II TA, essential genes for metabolism, housekeeping genes and any sequences exist in potential HGT receivers but NOT our coli can also be used. Even if the design cannot kill thieves, it can still weaken receivers and reduce the advantages a stolen antibiotic gene may bring in, thus reducing the degree of danger in HGT.</p> | |
| - | </ | + | |
| - | + | <p style="text-indent: 2em;">A final note: in the past, the repression efficiencies of antisense RNA in bacteria are low. However, after the invention of paired termini antisense RNA (PTasRNA) method, incorporation of U turn/YUNR motif, etc., the concept of competitive inhibition will become more and more feasible for the microbial synthetic biology community[[#Ref|[1 , ]][[#Ref|3 , ]][[#Ref|4] ]].</p> | |
| - | < | + | |
| - | </ | + | [[File:NTU-Taida-AsRNA.png|700px|center|thumb|alt=new idea about safety|Is it possible to use RNA interaction to avoid HGT? ]] |
| - | + | ||
| - | + | == Safety of Delivery System == | |
| - | </ | + | <p style="text-indent: 2em;">Many turn off strategies have been developed, most of them are inducible suicide systems that can be activated at certain conditions. In our project, we plan to use temperature as the activating signal (or alternatively, small molecules). When the course of treatment ends, leaving human body (temperature drop from 37C to room temperature) or administration of small amount of drugs (e.g. tetracycline) will cause suicide gene activation, thereby avoiding recombinant strain/gene pollution. And splitting suicide system to provide repression in trans can prevent plasmid transferring to wild type strains. Many off-the-rack parts can be used.</p> |
| - | + | ||
| + | [[File:NTU-Taida-Suicide-system.png|300px|thumb|left|alt=new idea about safety|general scheme of suicide system ]] | ||
| + | |||
| + | <p style="text-indent: 2em;">However, these designs cannot completely eliminate the risk of horizontal gene transfer (HGT), where recombinant genes can move to other organisms independent of suicide system. Therefore, in addition to suicide system, we propose a new concept to deal with HGT risks by RNA interaction[[#Ref|[1] ]]. Although bacteria lack RNAi pathway, expressing well designed antisense RNAs have been shown to have inhibitory effect on target RNAs through competitive inhibition.A recent study has demonstrated that expressing peptide nucleic acid (PNA) antisense to antitoxin RNA 5' sequence kills bacteria cells [[#Ref|[2] ]]. We reason that adding rationally designed antisense RNAs on the untranslated region of transcripts may interfere with target RNA’s function or translation, and we can employ this property to prevent HGT. For instance, HGT is more likely to occur between related species, such as lab E. coli & O157. Laboratory E. coli have inactivated all its hok/sok toxin-antitoxin system by mutation, but wild bacteria (especially pathogenic bacteria) usually have more active TA locus on its chromosome like E. coli O157. We plan to put a stem loop from hok mRNA, which can pair with sok RNA, 5’sequence on UTR of antibiotic resistance genes used in our PepdEx system. IF wild bacteria steal our antibiotic resistance genes and express it, its antitoxin will be competitive inhibited, and its toxin will get expressed to kill the thief. This prevents HGT between lab & wild coli. This concept can have wide extension. In addition to targeting antitoxin (functional RNA) of type I TA, it is also possible to design antisense sequences that target RBS to down-regulate targeted protein. Targeting antitoxin of type II TA, essential genes for metabolism, housekeeping genes and any sequences exist in potential HGT receivers but NOT our coli can also be used. Even if the design cannot kill thieves, it can still weaken receivers and reduce the advantages a stolen antibiotic gene may bring in, thus reducing the degree of danger in HGT.</p> | ||
| + | |||
| + | <p style="text-indent: 2em;">A final note: in the past, the repression efficiencies of antisense RNA in bacteria are low. However, after the invention of paired termini antisense RNA (PTasRNA) method, incorporation of U turn/YUNR motif, etc., the concept of competitive inhibition will become more and more feasible for the microbial synthetic biology community[[#Ref|[1 , ]][[#Ref|3 , ]][[#Ref|4] ]].</p> | ||
| + | |||
| + | [[File:NTU-Taida-AsRNA.png|700px|center|thumb|alt=new idea about safety|Is it possible to use RNA interaction to avoid HGT? ]] | ||
| - | </li> | + | ===Reference=== |
| + | <ol id="Ref"> | ||
| + | <li >Field, C.M. and D.K. Summers, Multicopy plasmid stability: revisiting the dimer catastrophe. Journal of Theoretical Biology, 2011. 291: p. 119-27.</li> | ||
| + | <li>Falvey, E. and N.D.F. Grindley, Contacts between Gamma-Delta-Resolvase and the Gamma-Delta-Res Site. Embo Journal, 1987. 6(3): p. 815-821.</li> | ||
| + | <li>Grindley, N.D.F., et al., Transposon-Mediated Site-Specific Recombination - Identification of 3 Binding-Sites for Resolvase at the Res-Sites of Gamma-Delta and Tn3. Cell, 1982. 30(1): p. 19-27.</li> | ||
| + | <li>Wells, R.G. and N.D.F. Grindley, Analysis of the Gamma-Delta Res Site - Sites Required for Site-Specific Recombination and Gene-Expression. Journal of Molecular Biology, 1984. 179(4): p. 667-687.</li> | ||
| + | <li>Guhathakurta, A., I. Viney, and D. Summers, Accessory proteins impose site selectivity during ColE1 dimer resolution. Molecular Microbiology, 1996. 20(3): p. 613-620.</li> | ||
| + | <li>Mullins, R.D., Bacterial Chromosome Segregation. Annu Rev Cell Dev Biol, 2009.</li> | ||
| + | <li>Godfrin-Estevenon, A.M., F. Pasta, and D. Lane, The parAB gene products of Pseudomonas putida exhibit partition activity in both P. putida and Escherichia coli. Molecular Microbiology, 2002. 43(1): p. 39-49.</li> | ||
| + | <li>Gerdes, K., et al., Mechanism of killer gene activation. Antisense RNA-dependent RNase III cleavage ensures rapid turn-over of the stable hok, srnB and pndA effector messenger RNAs. Journal of Molecular Biology, 1992. 226(3): p. 637-49.</li> | ||
| + | <li>Thisted, T. and K. Gerdes, Mechanism of post-segregational killing by the hok/sok system of plasmid R1. Sok antisense RNA regulates hok gene expression indirectly through the overlapping mok gene. Journal of Molecular Biology, 1992. 223(1): p. 41-54.</li> | ||
| + | <li>Szekeres, S., et al., Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Molecular Microbiology, 2007. 63(6): p. 1588-1605.</li> | ||
| + | <li>Faridani, O.R., et al., Competitive inhibition of natural antisense Sok-RNA interactions activates Hok-mediated cell killing in Escherichia coli. Nucleic Acids Research, 2006. 34(20): p. 5915-5922.</li> | ||
| + | <li>Gerdes, K. and E.G.H. Wagner, RNA antitoxins. Current Opinion in Microbiology, 2007. 10(2): p. 117-124.</li> | ||
| + | <li>Good, L. and J.E. Stach, Synthetic RNA silencing in bacteria - antimicrobial discovery and resistance breaking. Front Microbiol, 2011. 2: p. 185.</li> | ||
| + | <li>Faridani, O.R., et al., Competitive inhibition of natural antisense Sok-RNA interactions activates Hok-mediated cell killing in Escherichia coli. Nucleic Acids Research, 2006. 34(20): p. 5915-22.</li> | ||
| + | <li>Lucks, J.B., et al., Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc Natl Acad Sci U S A, 2011. 108(21): p. 8617-22.</li> | ||
| + | <li>Franch, T., et al., Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. Journal of Molecular Biology, 1999. 294(5): p. 1115-25.</li> | ||
</ol> | </ol> | ||
<!-- EOF --> | <!-- EOF --> | ||
{{:Team:NTU-Taida/Templates/ContentEnd}}{{:Team:NTU-Taida/Templates/Footer|ActiveNavbar=Project, #nav-Project-Stability}} | {{:Team:NTU-Taida/Templates/ContentEnd}}{{:Team:NTU-Taida/Templates/Footer|ActiveNavbar=Project, #nav-Project-Stability}} | ||
Revision as of 20:10, 25 October 2012
Stability
Every GM system that functions outside the laboratories will face two major problems: system stability and safety! Without selective pressure, we have to deal with plasmid instability; to make our ''E. coli'' colonized bowel we have to use recA+ strains which may cause plasmid multimer. Our genetically modified ''E. coli'' will also contact with many kinds of bacteria and is under the risk of horizontal gene transfer.The following segment is our struggle against these obstacles.
Contents |
Stability of Delivery System
Briefing
As our PepdEx system consists of several plasmids and will function outside of the laboratory (human gut) which lacks of antibiotic selection pressure, plasmid segregation stability is critical that determines whether every single E. Coli cell contains the original system with the designated function. Inspired by natural plasmid & mobile gene element, we cope with segregation instability by incorporating three modules, i.e. partition system, Multimer resolution system and toxin antitoxin system, on top of our system. We think that this layer of regulation will potentially benefit the entire synthetic biology community, as synthetic systems are becoming increasingly complex with fine controls (e.g. on low copy plasmids).
Obstacles
Plasmid instability problems come majorly from the segregational instability,the burden effect and the dimer catastrophe.
- Segregational instability
Plasmids are randomly (possibly uneven) distributed inside a bacterium. After cell division, one of the progenies may lose plasmids.
- Burden Effect
Cells bearing our PepdEx system have to spend extra energy/resource, so will not grow as good as plasmid-free cells. This metabolic burden will cause our coli to gradually lose pieces of circuits, and the growth rate difference will eventually eliminate plasmid-bearing bacteria in the population.
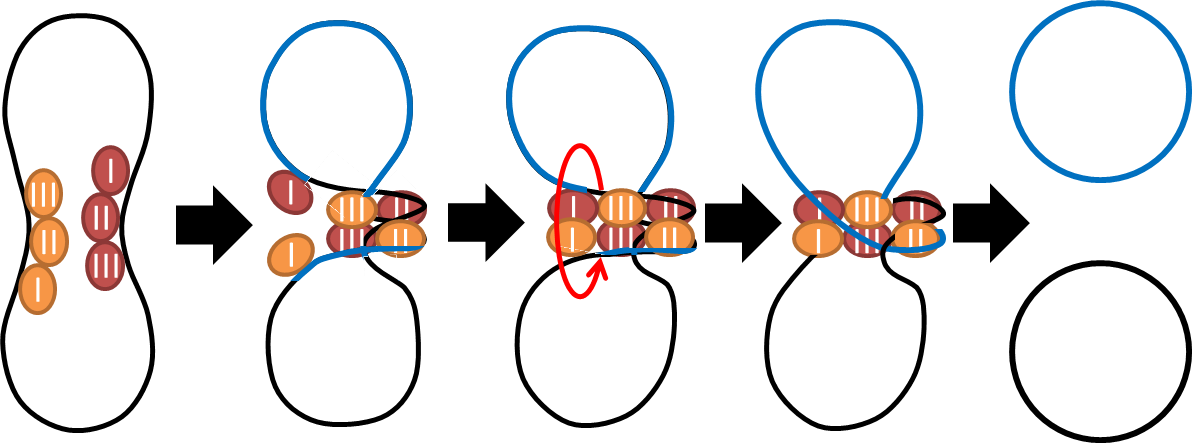
- Dimer Catastrophe
Homologues recombination may cause plasmid multimers which will increases segregational instability and burden[1] . This is the reason why most lab coli are recA1 mutants. But since our coli must be able to colonize in the gut, it should be recA+ wild type strain. That's the problem!
Stabilization Modules
1. Multimer Resolution System:Tn1000(gamma delta) resolution system
To deal with multimerization, we cloned an cassette (BBa_K817018) which encodes an autoregulated resolves (serine type recombinase) from E.coli F plasmid transposon tn1000 (tn3 family)[2] . Its promoter region consists of 3 sub-sites(res site) and can process recombination[3] . This cassette can resolve multimer that formed during replicative transposition[4] . It can also resolve plasmid multimer to increase plasmid stability by avoiding dimer catastrophe. Multimer resolution system (MRS) provides analogous functions to E.coli chromosome XerCD/dif but acts independent of cell cycle, DNA localization and may have higher efficiency on plasmids compared with slow XerCD system[5] .
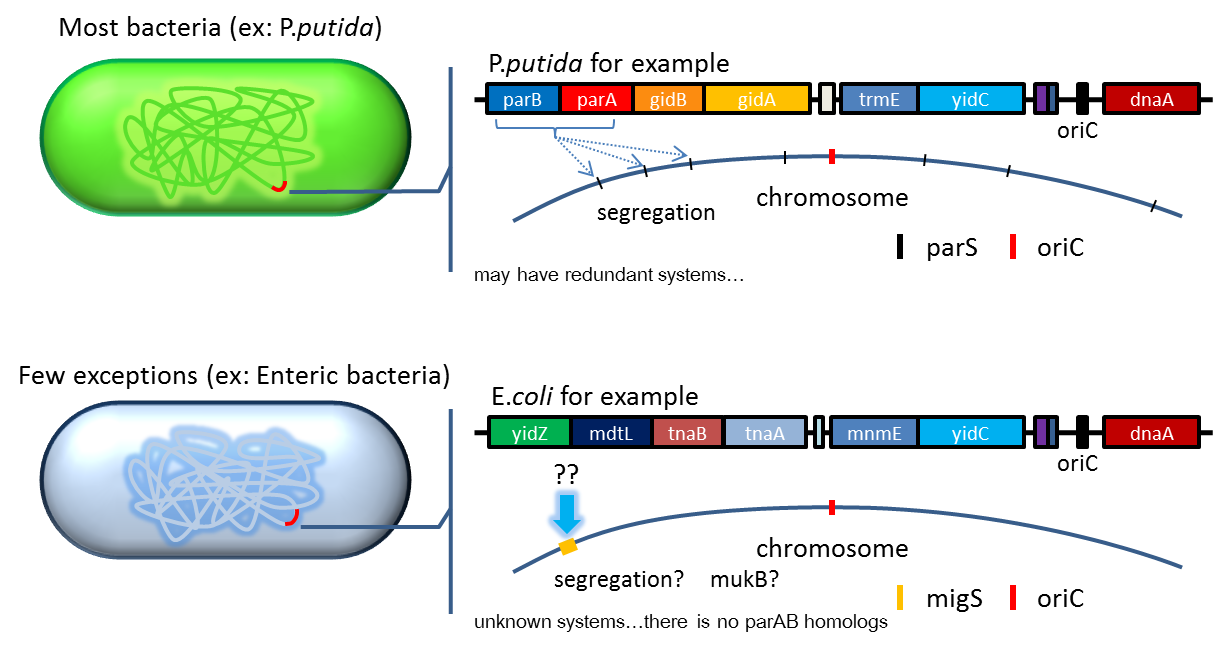
2. Partition System:from Pseudomonas putida KT2440
Bacterium distributes chromosomes/plasmids evenly to its progenies by a dynamic system that includes SMC like proteins, type Ia partition system and so on[6] . Type Ia partition system segregates chromosomes/plasmids in a process akin to Eukaryotic mitosis. It can be found on most eubacteria. However, E. coli is not the case.
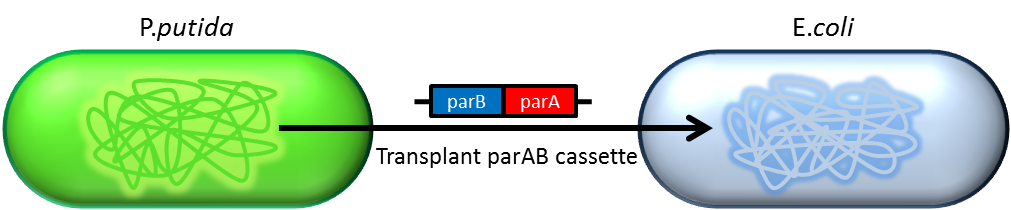
Previous study have shown that when providing parAB of P.putida in trans and carrying parS site in cis, segregation of low copy plasmids (mini F) can be stabilized in E. coli.[7] ]].The BioBrick vector pSB2K3 is a mini F plasmid, thus we expect we can stabilize its segregation this way. Our team have made a modified version of pSB2K3 with parS site (BBa_K817017), and plan to use it to harbor our PepdEx system (a low copy number plasmid allows us to fine tune our systems controls e.g. with smaller number of operator sites). We have also cloned the parAB operon (BBa_K817019), and provided the registry with the short 16bp parS site (BBa_K817016) for future applications.
3. Post-Segregation Killing:srnBC toxin-antitoxin system
No matter how well our partition system and multimer resolution system work, inevitably there will still be some bacteria losing plasmids (pieces of the circuit). These cells with incomplete system will not function as hoped. We sentence these cells to death to solve the problem.

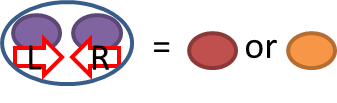
| 
|
Type I toxin-antitoxin srnBC is an ideal executor which belongs to hok/sok homologues. It expresses stable toxin encoding RNA and short-lived antitoxin RNA that can neutralize toxin RNA by RNA interaction and RNase III cleavage[9] . It acts as post segregation killing system – a bacterial cell will get killed if it loses the DNA (genomic islands, plasmids, mobile gene elements) that contains the module[10] . We plan to use it to reduce plasmid-lost cells in our coli population, in the hope to make applications without antibiotic selection more feasible. We have cloned the cassette with BioBrick standard (BBa_K817015). Following pictures show the mechanism (based on hok/sok).
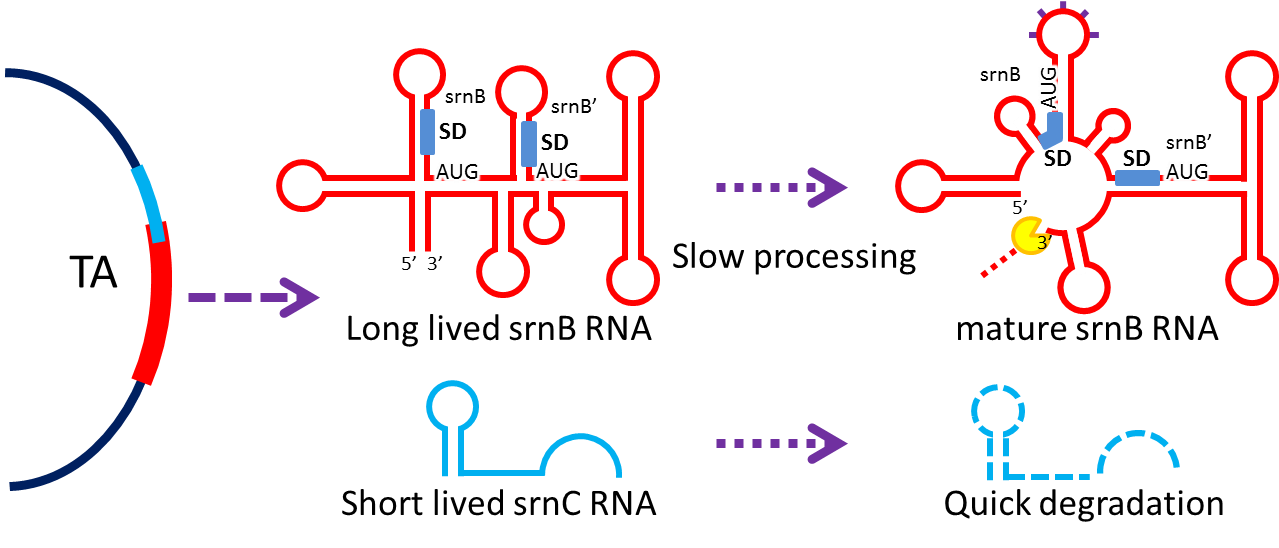
TA loci express two kinds of RNA, namely srnB and srnC, respectively. Initially, srnB is inactive and stable (half-life in the order of 30 minutes [11] ) and will gradually be processed to active form producing hydrophobic toxic protein if srnC RNA does not exist; srnC is relative unstable and is degraded quickly, and it overlaps and compliments to 5’sequence of srnB.
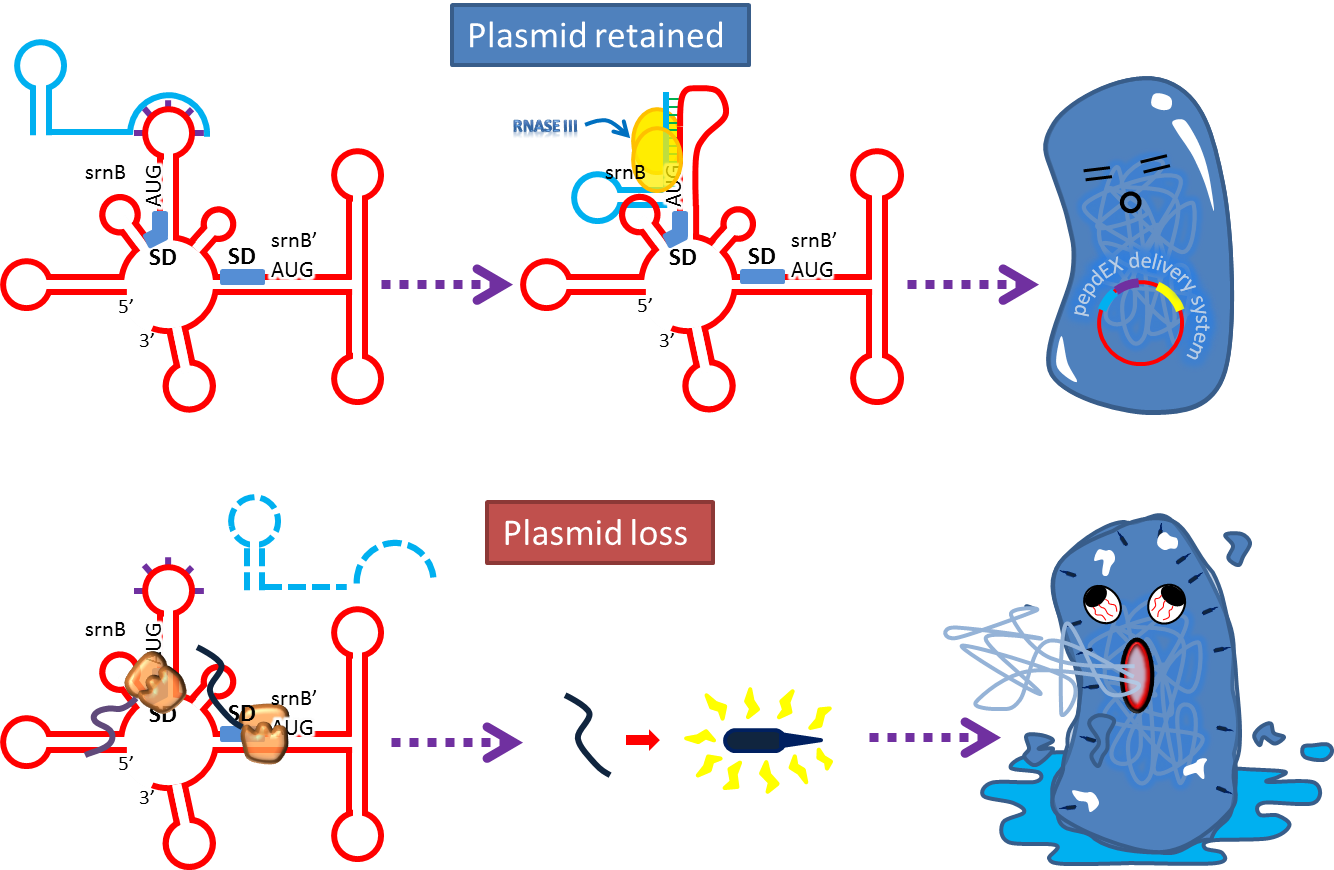
At normal condition (plasmid retained), srnC can pair with processed srnB and forms double stranded RNA that is susceptible to RNase III cleavage and thus prevents toxin production. If the plasmid loses, srnC will quickly disappear without replenishing and srnB will show its power[12] .
4. Reduce Burden Effect
Finally, beyond these modules, reducing the burden effect is in general important to the stability of complex artificial systems in an intact E. coli cell without antibiotic selection. We reason that a well designed system with a lower gene dosage will help.
High level gene expression and gene dosage leads to burden effect. Therefore, if two systems are encoded in different plasmids with the same overall performance, low copy plasmids are preferred over high copy ones. Again if with the same overall outcome, stabilizing mRNA is preferred over overexpressing the gene (unless for regulatory purposes). Place yourselves in E. coli's position! Reduce its metabolic burden as much as possible then it will work for you!
Safety of Delivery System
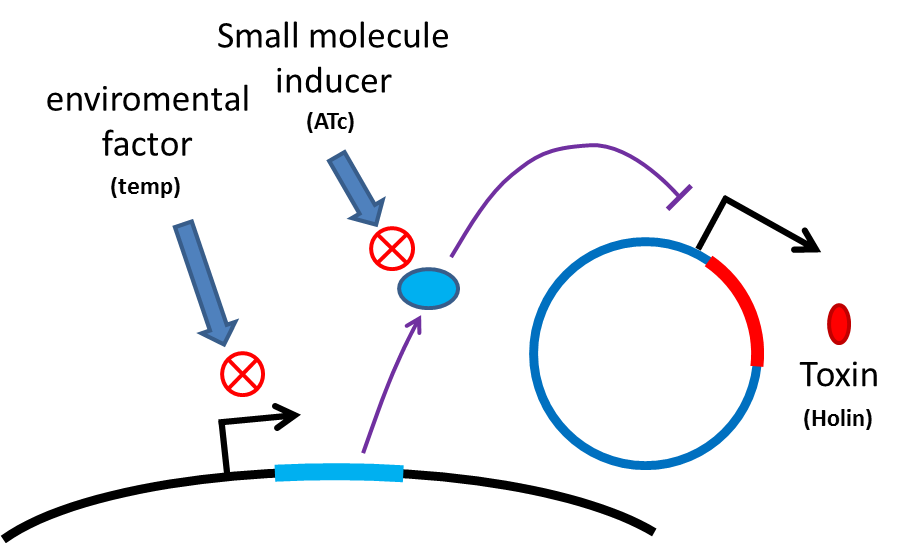
Many turn off strategies have been developed, most of them are inducible suicide systems that can be activated at certain conditions. In our project, we plan to use temperature as the activating signal (or alternatively, small molecules). When the course of treatment ends, leaving human body (temperature drop from 37C to room temperature) or administration of small amount of drugs (e.g. tetracycline) will cause suicide gene activation, thereby avoiding recombinant strain/gene pollution. And splitting suicide system to provide repression in trans can prevent plasmid transferring to wild type strains. Many off-the-rack parts can be used.
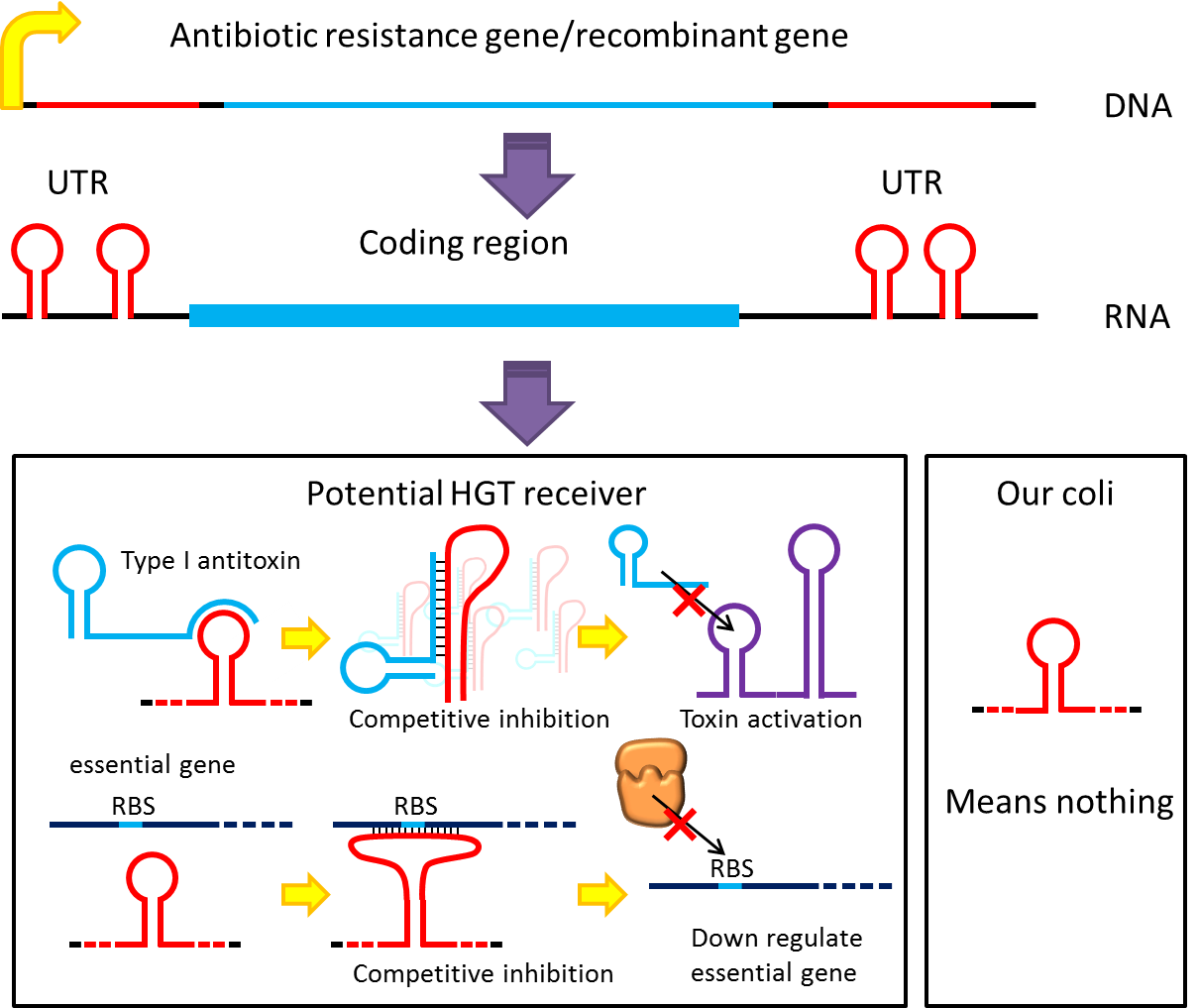
However, these designs cannot completely eliminate the risk of horizontal gene transfer (HGT), where recombinant genes can move to other organisms independent of suicide system. Therefore, in addition to suicide system, we propose a new concept to deal with HGT risks by RNA interaction[1] . Although bacteria lack RNAi pathway, expressing well designed antisense RNAs have been shown to have inhibitory effect on target RNAs through competitive inhibition.A recent study has demonstrated that expressing peptide nucleic acid (PNA) antisense to antitoxin RNA 5' sequence kills bacteria cells [2] . We reason that adding rationally designed antisense RNAs on the untranslated region of transcripts may interfere with target RNA’s function or translation, and we can employ this property to prevent HGT. For instance, HGT is more likely to occur between related species, such as lab E. coli & O157. Laboratory E. coli have inactivated all its hok/sok toxin-antitoxin system by mutation, but wild bacteria (especially pathogenic bacteria) usually have more active TA locus on its chromosome like E. coli O157. We plan to put a stem loop from hok mRNA, which can pair with sok RNA, 5’sequence on UTR of antibiotic resistance genes used in our PepdEx system. IF wild bacteria steal our antibiotic resistance genes and express it, its antitoxin will be competitive inhibited, and its toxin will get expressed to kill the thief. This prevents HGT between lab & wild coli. This concept can have wide extension. In addition to targeting antitoxin (functional RNA) of type I TA, it is also possible to design antisense sequences that target RBS to down-regulate targeted protein. Targeting antitoxin of type II TA, essential genes for metabolism, housekeeping genes and any sequences exist in potential HGT receivers but NOT our coli can also be used. Even if the design cannot kill thieves, it can still weaken receivers and reduce the advantages a stolen antibiotic gene may bring in, thus reducing the degree of danger in HGT.
A final note: in the past, the repression efficiencies of antisense RNA in bacteria are low. However, after the invention of paired termini antisense RNA (PTasRNA) method, incorporation of U turn/YUNR motif, etc., the concept of competitive inhibition will become more and more feasible for the microbial synthetic biology community[1 , 3 , 4] .
Safety of Delivery System
Many turn off strategies have been developed, most of them are inducible suicide systems that can be activated at certain conditions. In our project, we plan to use temperature as the activating signal (or alternatively, small molecules). When the course of treatment ends, leaving human body (temperature drop from 37C to room temperature) or administration of small amount of drugs (e.g. tetracycline) will cause suicide gene activation, thereby avoiding recombinant strain/gene pollution. And splitting suicide system to provide repression in trans can prevent plasmid transferring to wild type strains. Many off-the-rack parts can be used.
However, these designs cannot completely eliminate the risk of horizontal gene transfer (HGT), where recombinant genes can move to other organisms independent of suicide system. Therefore, in addition to suicide system, we propose a new concept to deal with HGT risks by RNA interaction[1] . Although bacteria lack RNAi pathway, expressing well designed antisense RNAs have been shown to have inhibitory effect on target RNAs through competitive inhibition.A recent study has demonstrated that expressing peptide nucleic acid (PNA) antisense to antitoxin RNA 5' sequence kills bacteria cells [2] . We reason that adding rationally designed antisense RNAs on the untranslated region of transcripts may interfere with target RNA’s function or translation, and we can employ this property to prevent HGT. For instance, HGT is more likely to occur between related species, such as lab E. coli & O157. Laboratory E. coli have inactivated all its hok/sok toxin-antitoxin system by mutation, but wild bacteria (especially pathogenic bacteria) usually have more active TA locus on its chromosome like E. coli O157. We plan to put a stem loop from hok mRNA, which can pair with sok RNA, 5’sequence on UTR of antibiotic resistance genes used in our PepdEx system. IF wild bacteria steal our antibiotic resistance genes and express it, its antitoxin will be competitive inhibited, and its toxin will get expressed to kill the thief. This prevents HGT between lab & wild coli. This concept can have wide extension. In addition to targeting antitoxin (functional RNA) of type I TA, it is also possible to design antisense sequences that target RBS to down-regulate targeted protein. Targeting antitoxin of type II TA, essential genes for metabolism, housekeeping genes and any sequences exist in potential HGT receivers but NOT our coli can also be used. Even if the design cannot kill thieves, it can still weaken receivers and reduce the advantages a stolen antibiotic gene may bring in, thus reducing the degree of danger in HGT.
A final note: in the past, the repression efficiencies of antisense RNA in bacteria are low. However, after the invention of paired termini antisense RNA (PTasRNA) method, incorporation of U turn/YUNR motif, etc., the concept of competitive inhibition will become more and more feasible for the microbial synthetic biology community[1 , 3 , 4] .
Reference
- Field, C.M. and D.K. Summers, Multicopy plasmid stability: revisiting the dimer catastrophe. Journal of Theoretical Biology, 2011. 291: p. 119-27.
- Falvey, E. and N.D.F. Grindley, Contacts between Gamma-Delta-Resolvase and the Gamma-Delta-Res Site. Embo Journal, 1987. 6(3): p. 815-821.
- Grindley, N.D.F., et al., Transposon-Mediated Site-Specific Recombination - Identification of 3 Binding-Sites for Resolvase at the Res-Sites of Gamma-Delta and Tn3. Cell, 1982. 30(1): p. 19-27.
- Wells, R.G. and N.D.F. Grindley, Analysis of the Gamma-Delta Res Site - Sites Required for Site-Specific Recombination and Gene-Expression. Journal of Molecular Biology, 1984. 179(4): p. 667-687.
- Guhathakurta, A., I. Viney, and D. Summers, Accessory proteins impose site selectivity during ColE1 dimer resolution. Molecular Microbiology, 1996. 20(3): p. 613-620.
- Mullins, R.D., Bacterial Chromosome Segregation. Annu Rev Cell Dev Biol, 2009.
- Godfrin-Estevenon, A.M., F. Pasta, and D. Lane, The parAB gene products of Pseudomonas putida exhibit partition activity in both P. putida and Escherichia coli. Molecular Microbiology, 2002. 43(1): p. 39-49.
- Gerdes, K., et al., Mechanism of killer gene activation. Antisense RNA-dependent RNase III cleavage ensures rapid turn-over of the stable hok, srnB and pndA effector messenger RNAs. Journal of Molecular Biology, 1992. 226(3): p. 637-49.
- Thisted, T. and K. Gerdes, Mechanism of post-segregational killing by the hok/sok system of plasmid R1. Sok antisense RNA regulates hok gene expression indirectly through the overlapping mok gene. Journal of Molecular Biology, 1992. 223(1): p. 41-54.
- Szekeres, S., et al., Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Molecular Microbiology, 2007. 63(6): p. 1588-1605.
- Faridani, O.R., et al., Competitive inhibition of natural antisense Sok-RNA interactions activates Hok-mediated cell killing in Escherichia coli. Nucleic Acids Research, 2006. 34(20): p. 5915-5922.
- Gerdes, K. and E.G.H. Wagner, RNA antitoxins. Current Opinion in Microbiology, 2007. 10(2): p. 117-124.
- Good, L. and J.E. Stach, Synthetic RNA silencing in bacteria - antimicrobial discovery and resistance breaking. Front Microbiol, 2011. 2: p. 185.
- Faridani, O.R., et al., Competitive inhibition of natural antisense Sok-RNA interactions activates Hok-mediated cell killing in Escherichia coli. Nucleic Acids Research, 2006. 34(20): p. 5915-22.
- Lucks, J.B., et al., Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc Natl Acad Sci U S A, 2011. 108(21): p. 8617-22.
- Franch, T., et al., Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. Journal of Molecular Biology, 1999. 294(5): p. 1115-25.
 "
"