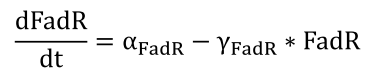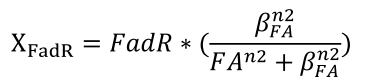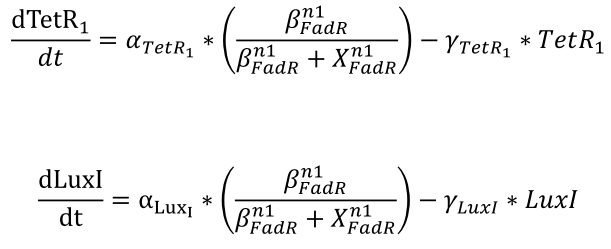Team:NTU-Taida/Modeling/Single-Cell
From 2012.igem.org
| Line 1: | Line 1: | ||
{{:Team:NTU-Taida/Templates/Header}}{{:Team:NTU-Taida/Templates/Navbar}}{{:Team:NTU-Taida/Templates/Sidebar}}{{:Team:NTU-Taida/Templates/ContentStart}} | {{:Team:NTU-Taida/Templates/Header}}{{:Team:NTU-Taida/Templates/Navbar}}{{:Team:NTU-Taida/Templates/Sidebar}}{{:Team:NTU-Taida/Templates/ContentStart}} | ||
| - | {{:Team:NTU-Taida/Templates/BSHero|Title= | + | {{:Team:NTU-Taida/Templates/BSHero|Title=Single Cell Model|Content=<p></p>}} |
| + | ==Introduction== | ||
| + | ===Motivation=== | ||
| + | Inferring from our circuit design, we expect to see an anti-noise response of GLP1 in spite of the fluctuating level of fatty acid. However, since the overall response of GLP1 strongly depends on the actual dynamics and equilibrium state of the species involved on the network, we are not able to confirm the anti-noise function without a detailed, quantitative analysis. Furthermore, the overall sensing threshold of fatty acid in our circuit plays a critical role in the function of our system, determining if our system is able to distinguish between the basal fatty acid level (the noise) and the FA level after food intake (the signal) and respond properly. Therefore, we performed single cell modeling to confirm the function of our circuit and examine the threshold of our high pass filter. | ||
| + | ===Overview=== | ||
| + | The Single cell model describes the species involved in our synthetic circuit of a single pepdex E. coli. By non-linear ordinary differential equations (ODEs), the change in species concentrations over time is simulated, with most of the ODEs following Hill kinetics. The parameters used in our single cell model are derived from studies supported by experimental evidence, as listed in our Parameters part. After thorough analysis, some of the parameters are adjusted to meet our own experimental approaches, as described separately in the following sections. Our single cell model was simulated using numerical Matlab solvers. | ||
| - | {{:Team:NTU-Taida/Templates/ContentEnd}}{{:Team:NTU-Taida/Templates/Footer|ActiveNavbar= | + | ===Our Model=== |
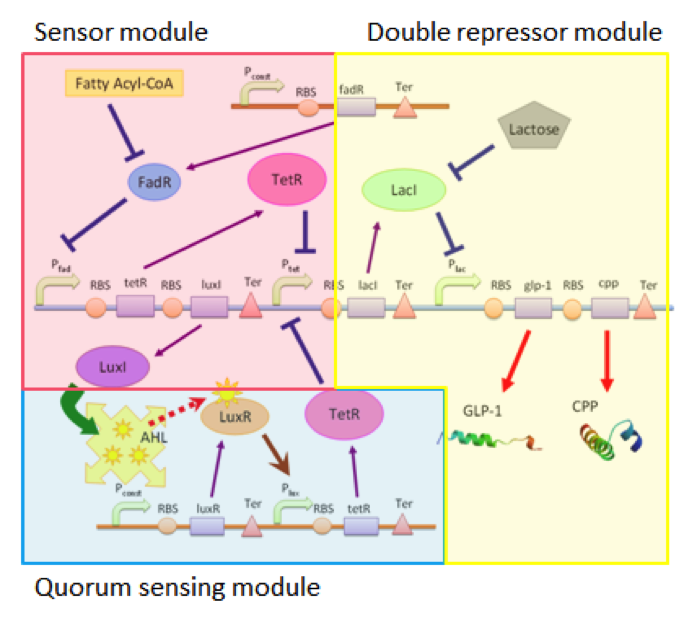
| + | Our circuit can be divided into three connected but independent modules: the sensor module, double repressor module and the quorum sensing module, as shown in figure 1. | ||
| + | |||
| + | [[FIle:NTU-Taida-Model-Single-fig1.png|450px|thumb|center|structure of MRS]] | ||
| + | |||
| + | |||
| + | The three modules are almost independent, connected between each other only by a single species. The independence of the three modules enables us to adjust each module according to different application without affecting the function of other modules. For example, we can use different sensors with the double repressor and the quorum sensing modules, allowing our cells to act as biological factories able to respond to different substances if the need arises. | ||
| + | |||
| + | ==Sensing Module== | ||
| + | |||
| + | Our sensing module senses the presence of fatty acid by the function of the ''fadR'' promoter. Fatty acids bind to FadR proteins and block the repression activity of FadR to the ''fadR'' promoter, therefore enabling the expression of the genes regulated by ''pfadR''. In this module, we simulate the sensor by separating the chemical pathway into three consecutive steps : (a.) the expression of protein FadR (b.) the binding of FA to FadR proteins, and (c. ) the repression of ''pfadR'' by FadR proteins. | ||
| + | |||
| + | <html><ol style="list-style-type: lower-latin;"> | ||
| + | <li>The expression of protein FadR is simply controlled by a constitutively expressed promoter, and therefore is governed by a simple equation</html><br />[[FIle:NTU-Taida-Single-eq1.png|center|structure of MRS]]<html></li> | ||
| + | |||
| + | <li>We describe the binding of FAs to FadR proteins by an equation derived from Michaelis-Menten kinetics, which determines the amount of effective FadR repressor, defined as X<sub>FadR</sub></html><br />[[FIle:NTU-Taida-Single-eq2.png|center|structure of MRS]]<html></li> | ||
| + | |||
| + | <li>The effective repressors follow Hill kinetics to inhibit the expression of the genes regulated by pfadR, which determines the resulting concentration of TetR1 and LuxI.</html><br />[[FIle:NTU-Taida-Single-eq3.png|center|structure of MRS]]<html></li> | ||
| + | </ol></html> | ||
| + | |||
| + | {{:Team:NTU-Taida/Templates/ContentEnd}}{{:Team:NTU-Taida/Templates/Footer|ActiveNavbar=Modeling, #nav-Modeling-Single}} | ||
Revision as of 12:44, 26 September 2012
Single Cell Model
Contents |
Introduction
Motivation
Inferring from our circuit design, we expect to see an anti-noise response of GLP1 in spite of the fluctuating level of fatty acid. However, since the overall response of GLP1 strongly depends on the actual dynamics and equilibrium state of the species involved on the network, we are not able to confirm the anti-noise function without a detailed, quantitative analysis. Furthermore, the overall sensing threshold of fatty acid in our circuit plays a critical role in the function of our system, determining if our system is able to distinguish between the basal fatty acid level (the noise) and the FA level after food intake (the signal) and respond properly. Therefore, we performed single cell modeling to confirm the function of our circuit and examine the threshold of our high pass filter.
Overview
The Single cell model describes the species involved in our synthetic circuit of a single pepdex E. coli. By non-linear ordinary differential equations (ODEs), the change in species concentrations over time is simulated, with most of the ODEs following Hill kinetics. The parameters used in our single cell model are derived from studies supported by experimental evidence, as listed in our Parameters part. After thorough analysis, some of the parameters are adjusted to meet our own experimental approaches, as described separately in the following sections. Our single cell model was simulated using numerical Matlab solvers.
Our Model
Our circuit can be divided into three connected but independent modules: the sensor module, double repressor module and the quorum sensing module, as shown in figure 1.
The three modules are almost independent, connected between each other only by a single species. The independence of the three modules enables us to adjust each module according to different application without affecting the function of other modules. For example, we can use different sensors with the double repressor and the quorum sensing modules, allowing our cells to act as biological factories able to respond to different substances if the need arises.
Sensing Module
Our sensing module senses the presence of fatty acid by the function of the fadR promoter. Fatty acids bind to FadR proteins and block the repression activity of FadR to the fadR promoter, therefore enabling the expression of the genes regulated by pfadR. In this module, we simulate the sensor by separating the chemical pathway into three consecutive steps : (a.) the expression of protein FadR (b.) the binding of FA to FadR proteins, and (c. ) the repression of pfadR by FadR proteins.
- The expression of protein FadR is simply controlled by a constitutively expressed promoter, and therefore is governed by a simple equation
- We describe the binding of FAs to FadR proteins by an equation derived from Michaelis-Menten kinetics, which determines the amount of effective FadR repressor, defined as XFadR
- The effective repressors follow Hill kinetics to inhibit the expression of the genes regulated by pfadR, which determines the resulting concentration of TetR1 and LuxI.
 "
"