Team:NRP-UEA-Norwich/Week4
From 2012.igem.org
Khadijaouadi (Talk | contribs) |
Khadijaouadi (Talk | contribs) |
||
| (17 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
=Week 4= | =Week 4= | ||
| - | This week we made great progress into designing our own constructs for the comparative circuit. The amount of lab work | + | This week we made great progress into designing our own constructs for the comparative circuit. Many challenges in the design of the complementary sequences were overcome such as the creation of scars and the standard we would design it for. The amount of lab work had definitely picked up. We are extremely close to creating our own first BioBricks. Hopefully by next week, we will have them. |
| - | ==Day 1== | + | ==Day 1 (30/07/12)== |
===Research and Meetings=== | ===Research and Meetings=== | ||
| - | Khadija and Pascoe | + | Khadija and Pascoe encountered obstacles with stop codons over the weekend and two possible solutions were thought up. One was a scaffold construction, the other one the assembly using standard 23. The team leaned towards the assembly 23 standard as it's less risky than the scaffold, however was decided that further research is needed in order to make an informed decision. |
| + | Two main constructs were developed; one saw overlapping stands, the other saw strands together. Khadija produced a logic table of codons that could bind to one another. Amy also came to visit in order to look at the studio we bfor our film being recorded on Wednesday. | ||
===Labs=== | ===Labs=== | ||
| - | + | Joy and Lukas continued planning the ligation of BM/MB promoter into the pSB1C3 backbone which will give us our first two BioBricks. As the B-M fragment with the correct prefix and suffix endings was ready from last week, the [https://2012.igem.org/Team:NRP-UEA-Norwich/Protocol#Ligation iGEM protocol] was performed. | |
| - | + | Following the ligation, Rebecca transformed the potential BioBrick into Bioline competent alpha cells and the cell cultures were grown on the appropriate media (now containing 100ug/µl of chloramphenicol) at 37 °C over night. | |
| + | Observing cells transformed on Friday (that were stored in the fridge after the initial incubation for 24 hours), Russell and Rachel found that the cultures had not grown as much as they would have liked. Therefore the plates were returned to the 37 °C incubator to grow some more overnight. | ||
| - | + | ==Day 2 (31/07/12)== | |
| - | + | ||
| - | ==Day 2== | + | |
===Research=== | ===Research=== | ||
| - | + | Russell and Rachel researched nitrite reductases (NOSs) and their sequences, to be able to determine it functionality as a reporter in the NO sensing gene construct and its ability to raise NO levels to toxic concentrations. In addition the sequences were observed to judge whether it would be feasible to get them synthesised. | |
| - | + | The whole team worked together with Khadija and Pascoe on the complementary DNA sequences for the comparative circuit; four members helping with Khadija's logic map and three with Pascoe's idea. | |
| - | . The whole team worked with Khadija and Pascoe on the DNA | + | |
===Labs=== | ===Labs=== | ||
| - | + | As the colonies transformed with the ligation product of B-M in pSB1C3 from yesterday showed no growth, Rebecca redid transformation using the same ligation product and incorporated untransformed alpha cells as a negative control and chloramphenicol resistant PyeaR cells as a positive controls. | |
| + | |||
| + | More chloramphenicol plates were poured to stock pile for the transformation of ligation products in the coming weeks. The transformants from yesterday were incubated for longer in hope that colonial growth would appear. | ||
| - | + | Lukas and Joy isolated plasmid DNA from "E.coli" containing the hybrid promoter MB plasmid. They then double digested the DNA using EcoRI and PstI to separate the MB promoter with iGEM prefix and suffix attached. Double digests with SacI and SpeI of the same plasmid were also performed as validation of the DNA. Both digests were run on an agarose gel (''Figure 1.''). [[File:M-B_in_pUC57_E+P_S2.png | thumb | '' '''Figure 1.''' Gel electrophoresis of B-M in pUC57, double digested with EcoRI and PstI as well as validated with SacI and SpeI'']] | |
| + | The protocols can be seen [https://2012.igem.org/Team:NRP-UEA-Norwich/Protocol#Gel_Electrophoresis here.] | ||
| + | The validation digest was also carried out on the BM plasmid, that had been used in the ligation on Monday, to exclude the quality of the starting material of the transformation as a source of error. | ||
| + | After incubating the ''E. coli'' cultures, that had been transformed with RFP, eCFP and AraC promoter, for longer there was some success as more colonies were oberved on almost all the plates. Russell and Rachel inoculated 5 ml tubes of LB media (+ 5µl ampicillin) with transformed ''E. coli'' as followed: | ||
| - | + | Two tubes of RFP-transformed ''E. coli'' from different colonies on the 200µl plate | |
| - | + | Two tubes of eCFP-transformed ''E. coli'' from different colonies on the 200µl plate | |
| - | + | Four tubes of AraC Promoter-transformed ''E. coli'' from different colonies on the 200µl plate (due to the fact that promoters were needed for each of the fluorescent proteins, we decided to create double the amount). | |
| - | + | The inoculated media was placed in the 37 °C agitator and left overnight to grow. The protocol for this can be found [https://2012.igem.org/Team:NRP-UEA-Norwich/Protocol#Inoculations Here] | |
| - | + | ==Day 3 (01/08/12)== | |
| - | + | ||
| - | ==Day 3== | + | |
[[File:CombinedMRNA.png | thumb | The JK-R5-PL Construct]] | [[File:CombinedMRNA.png | thumb | The JK-R5-PL Construct]] | ||
| - | |||
| - | |||
===Labs=== | ===Labs=== | ||
| - | + | Rachel and Russell checked the inoculated media from the day before and observed that bacterial growth had not yielded enough cells to perform a likely successful experiments. Therefore it was decided to continue the incubation before attempting a DNA isolation via mini prep. | |
| - | . | + | Repeat transformations of BM/MB in pSB1C3 did not yield any viable cells. But the negative control showed no growth and the positive controlled did, from which it was concluded that the error must lie with the ligation and not with the transformation. |
| - | + | Rebecca purified the BM insert (cut from pUC57 with EcoRI and PstI, see ''Figure 2.''), cut out from the agarose gel slice and frozen the previous night. This was performed using a Promega gel purification kit to minimise the error that might be introduced with the Bioline kit. The protocol can be found on the [https://2012.igem.org/Team:NRP-UEA-Norwich/Protocol lab protocol page]. The isolated sample was run on an agarose gel. [[File:Bm_dd,_E+P,_Sx2.png | thumb | '' '''Figure 2.''' B-M in pUC57, double digested with EcoRI and PstI and validated with SpeI and SacI'']] | |
| - | |||
===Research=== | ===Research=== | ||
| - | In the morning Khadija, Pascoe, Rebecca and Russell looked at one of the proposed constructs and worked | + | In the morning Khadija, Pascoe, Rebecca and Russell looked at one of the proposed constructs and worked on producing a DNA sequence that forms two complimentary strands that would bind together once transcribed to mRNA. After a few hours of design and lots of adjustments and edits, we finally had our construct which we named the JK-R5-PL construct (using the initials of the team members). The sequence is the coming-together of two separate constructs, which would be in between the promoter and reporter sequences of the gene we were producing. We were pleased with the JK-R5-PL construct because it meant the ribosome binding site of both original constructs were covered, resulting in the two reporter proteins not to be expressed. This would ultimately result in the first step of the comparator circuit being completed! |
===Video=== | ===Video=== | ||
| - | Amy Congdon visited along with two of her colleagues to film for the final presentation video. The artist team had produced some fantastic visual adaptations of cancer cells and the bacteria | + | Amy Congdon visited along with two of her colleagues to film for the final presentation video. The artist team had produced some fantastic visual adaptations of cancer cells and the bacteria interacting with them while analysing the environment and act upon the information received. We also filmed the more concept-based aspect of the video, which looks at a future world where comparator circuit-containing bacteria are commonplace in all human beings, assessing the body for disease and reporting the results back to the person. |
| - | |||
| - | + | ==Day 4 (02/08/12)== | |
| - | + | WE RECEIVED A LETTER FROM DAVID ATTENBOROUGH!!! | |
| - | . | + | [[File:Letter.jpg | thumb | Woooooooo!!!]] |
| - | + | Posters for our human outreach event at the Forum were returned from the print shop and we were getting ready to start putting them up. | |
| - | + | ===Labs=== | |
| - | + | As the gel purification of MB/BM fragments did not yield DNA samples of high concentrations and we suspected that this might be the cause of the failed ligations, Joy performed a PCR on the MB/BM sequences in pUC57 to increase the amount of DNA we had at hand. The PCR was performed with a negative control and it is important to note that the primers used were standard amplification primers for the pUC57 backbone which allowed us to amplify the hybrid promoters as well as the iGEM suffix and prefix. | |
| - | + | Russell and Rachel proceeded with DNA isolations of the cells cultures containing the RFP, eCFP and AraC promoter BioBricks. The samples were put on ice for validation gel electrophoresis the next day. A Promega plasmid isolation kit was used and for each original cell culture, a small amount of unused cells was stored in the fridge as a back-up in case of any problems with the plasmid isolation. | |
| - | + | ==Day 5 (03/08/12)== | |
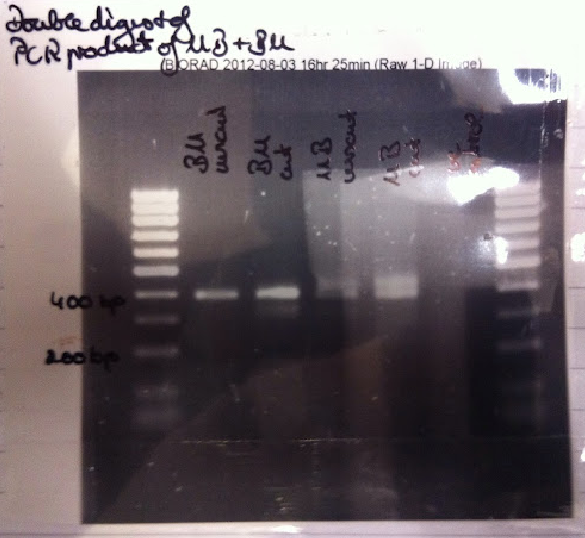
| - | + | To check DNA concentrations of BM and MB PCR products, nano drops were performed. We found that the DNA concentrations off all products were above 300ng/µl. To purify the samples and prevent primers, salt and buffers from interfering with downstream applications of the MB/BM genes, a Promega PCR cleanup kit was used. [[File:PCR_double_digest.png | thumb | '' '''Figure 3.''' PCR product of B-M/M-B in pUC57, digested using BanHI'']] | |
| - | + | Lukas carried out a double (EcoRI + PstI) and single (EcoRV) restriction digest on PyeaR + GFP (BBa_K381001). The single digest was used to validate whether the isolated plasmid DNA is in fact BBa_k381001. The double digest would yield a linearised pSB1C backbone and fragments of the PyeaR + GFP BioBrick. They will be utilised as a positive control in ligations and transformations. Gel electrophoresis was used to visualise the DNA fragements. The single digest was successful linearising the plasmid and thus confirming that the DNA, contained in the samples, was in fact PyeaR + GFP. The double digest was not successful and needed to be repeated. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | . Gel electrophoresis was used to | + | |
| - | . The | + | A double (EcoRI + PstI) and single (BamHI) digest was carried out on the PCR amplification of BM/MB. The enzymes EcoRI and PstI will release the promoter sequence from the primer sequences leaving them with sticky ends of the IGEM prefix and suffix. The single digest was used for validation. |
| + | Following the restriction digest, the DNA was nanodropped to check the concentrations and then run on a 1.4% agarose gel. If the PCR cloning, purification and digestion were successful, 251bp fragments should be produced. The gel showed shadowing and was not completely clear. The gel will have to be rerun. | ||
| + | The BamHI digested products were run on a gel. It showed 200bp fragments, suggesting that the insert was produced (''Figure 3.''). | ||
Latest revision as of 00:32, 27 September 2012
 "
"