Team:NRP-UEA-Norwich/Experiments
From 2012.igem.org
Welcome to the NRP UEA iGEM 2012 Wiki Lab Book
Please choose the relevant link to access our diary of that week!
Week Zero | Week One | Week Two | Week Three | Week Four | Week Five | Week Six | Week Seven | Week Eight | Week Nine | Week Ten | Week Eleven | Lab Protocols | Experiments
The following experiments, as the title suggests, are the experiments carried out to characterise both our BioBricks and others. In total we characterised PyeaR, BM and MB as well as gained experience in the use of CFP and RFP. The experiments written below are not in chronological order as we received the BioBricks at different points of our project. Given more time, there are further characterisation studies we would like to carry out. These are also described below.
The hybrid promoters, BM and MB were created to increase the flexibility of chassis a promoter can be used in. To fully characterise the functionality of BM and MB, it was therefore important that it can in fact work within mammalian and bacterial chassis. Through transformations and successful growth in potassium nitrate, it has been shown that BM and MB both function in bacterial cells.
To incorporate BM and MB into a mammalian system, transformations were not possible. Instead a transfection was carried out. Unlike the growth studies, where growing colonies were sufficient to prove that the DNA within plasmids were incorporated, reporter proteins (CFP and RFP) were attached to allow visual characterisation.
To transfect the cells we used lipid based transfection. To do this, media, transfection reagent, nitric oxide donor, DNA and cells were required. Below is the full list of reagents used:
. DMEM (Dulbecco’s Modified Eagle Medium) without serum to transfect and later treated with DMEM with serum and 10% Fetal Calf Serum. Cells are transfected without serum as serum interferes with the process but cells can die without serum so later is treated with serum.
. M-B +CFP was the DNA used. More than one sample was used. From nanodropping the concentrations of DNA were found to be both 500nm/µL. To transfect, 6.5 µL of DNA was used. The exact DNA we used was labelled MB2-C11a and MB2-C12a. The DNA was obtained from different colonies from the same plate.
. SNAP (S-Nitroso-N-Acetylpenicillamine) at a final concentration of 500µM. SNAP is the nitric oxide donor. Unlike bacterial cells where potassium nitrate could be used as a direct source. SNAP is metabolised by cells to produce NO which then induces the BioSensor.
. LipoD293 which is the transfection agent. This creates a membrane around the DNA which then binds to the mammalian cell and allows entrance of DNA into cell. This is much like endocytosis.
. The chassis used was MCF7 which is a human breast cancer cell line. The cells (30 µL) were seeded into a 6 channel slide at a concentration of 3 x 105 cells/ml.
In the process of transfection, LipoD293 was mixed with DNA at left for 15mins. Meanwhile, the media was removed from the cells and washed with serum free DMEM (100 µL). The transfection mixture was then added to the relevant channels on the slide. Below is how our channels were labelled.
For the full transfection protocol please click here
After the transfection, the media was changed to media containing serum and also SNAP was added. The cells were then viewed and the following images were obtained.
From the images, there are globular shapes which appear brighter on the outside than on the inside. We feel that there are a number of possibilities of what these may be. We feel that the more likely possibility is that these are dead cells. Another possibility is that these are vesicles formed after apoptosis.
We feel that these are more likely to be dead cells for a number of reasons. The main reason being that after the addition of SNAP, the larger cells show stress fibers on closer inspection, showing that these are cells which are not 'happy' with their environment. Thus, it can be assumed, that many will die. Furthermore, there are these globular shapes in the image of untransfected cells in SNAP and also in the image of transfected cells without SNAP. Therefore, this is a regular occurrence. Cell death also being a regular occurrence.
Another possibility is that NO has induced the cells to apoptose and this has lead to vesicles forming containing the fluorescent proteins (Yu, et al., 1999). In non transfected cells, there is less fluorescent proteins compared to the transfected cells and hence there are more fluorescing vesicles.
Following transfection, to test the cytotoxicity of NO, the number of cells after addition of SNAP was calculated. The MCF7 cells were seeded into 6 well plates again at a concentration of 2.5 x 105 cells/ml. SNAP was then added at 500µM 2 days after plating. The cells were counted 24 hours after the addition of SNAP. For the full cell count protocol please click here.
From previous studies such as that by Lala and Chakraborty, 2001 have shown that NO can lead to cytostasis and apoptosis. Our assay further confirmed this.
References:
Lala, P.K. and Chakraborty, C. (2001) 'Role of nitricoxide in carcinogenesis and tumour progression', The Lancelet, 2;149–156.
Yu, W., Simmons-Menchaca, M., Gapor, A., Sanders, B.G. and Kline, K. (1999) 'Induction of Apoptosis in Human Breast Cancer Cells by Tocopherols and Tocotrienols', Nutrition and Cancer, 33;26-32.
Three tubes of media were inoculated with E. coli transformed by the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K774005 B-M + RFP] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K774006 M-B + eCFP] BioBrick. Each tube then had potassium nitrate added to it at different concentrations; 0 mM, 1 mM and 10 mM respectively. The E. coli were grown over night and then spun down, fixed in 4% PFA and re-suspened in 500ul PBS. The samples were then analysed in an Acuri C6 or BD Aria II flow cytometer.
Note: This was the first time the flow cytometers at the University of East Anglia had been used with E. coli
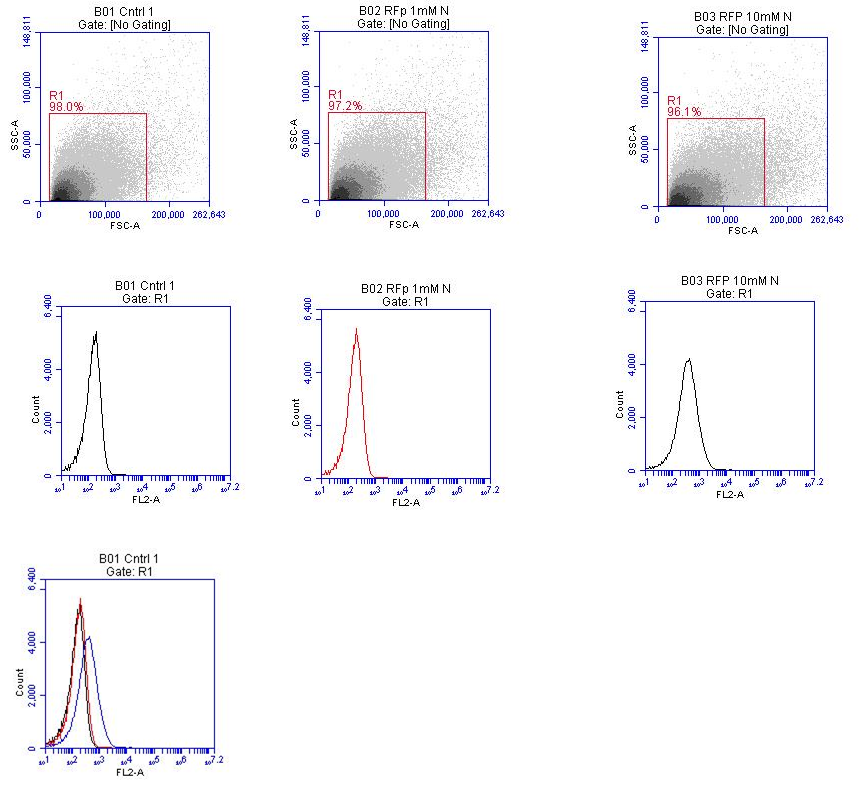
In this figure the image on the lower left of the fluorescence profiles overlayed on one another suggests that there is a slight difference in fluorescence intensity between the samples grown in media containing 0 mM potassium nitrate and 1 mM potassium nitrate, however there is a very noticeable difference in fluorescence between samples grown in 1 mM potassium nitrate and 10 mM potassium nitrate.
The data in this figure appears to corroborate with the data in Figure 13, showing a small difference in fluorescence between 0 mM potassium nitrate and 1 mM potassium nitrate, and then a much larger difference in fluorescence intensity following induction by 10 mM potassium nitrate.
The data in this figure suggests a slight increase in fluorescence intensity from 0 mM to 1 mM, and then a higher increase in fluorescence intensity from 1 mM to 10 mM.
The data in this figure corroborates with the data in figure 15 in suggesting a slight increase in fluorescence intensity from 0 mM to 1 mM, and then a higher increase in fluorescence intensity from 1 mM to 10 mM.
The effect of different potassium nitrate concentrations in growing media on the fluorescence activity of cells containing PyeaR + GFP was investigated. The experiment was meant as an initial qualitative test of the function of the BioBrick and to collect information on the influence of potassium nitrate as well as the development of a protocol for later studies on Bm and MB.
Culture of PyeaR +GFP containing cells were grown up in LB media of different potassium nitrate concentrations. The concentrations ranged from o mM to 40mM in 5 mM steps. After an incubation period of 24 hours at 37 °C, the cultures were pelleted and observed under UV light. The results can be observed below.
Following the experiment measuring the affect on the intensity of GFP fluorescence of PyeaR-GFP at different concentrations of potassium nitrate, a study on how different levels of potassium nitrate affect fluorescence intensity of BM and MB attached to fluorescent proteins was tested. Where previously the study was carried out in a qualitative fashion, this was carried out to collect data quantitatively using a fluorometer. Overnight cultures of BM and MB attached to RFPs and CFPs containing different levels of potassium nitrate prepared. Using 5ml of the cultures, cells were peletted. The overnight cultures contained different concentrations of potassium nitrate (0mM, 5mM, 10mM, 15mM and 20mM). These cells were then resuspended and lysed (full protocol on protocol page). These cultures were then diluted using Tris buffer (0.5ml sample to 1.5ml buffer). Readings from the fluorometer wavescans were recorded.
Comparing BM and MB with CFP fluorescence showed that BM-CFP intensity is relatively low when compared to MB. MB seems more sensitive to nitrate levels than BM. However, for both, it seems that once a threshold of nitrate levels is reached, there is not additional expression. Beyond this concentration, the intensity drops. This may be due to a number of factors. The cells may die due to overload of proteins which may have a toxic affect. For BM this threshold seems to be just above 20mM and for MB this appears to be around 15mM. This can be seen in the figure below.
Comparing BM and MB with RFP fluorescence showed a similar result to the CFP study. However both BM and MB have similar sensitivities to potassium nitrate levels, BM seems to be more sensitive to potassium nitrate concentration this time. From this study it is clear that beyond a certain concentration of nitrates, the fluorescence intensity stops increasing and decreases instead. Again, this may be due to the capacity or tolerance of the cell to hold the volume of fluorescence. protein.
The study involved testing the affects of transforming E.coli with different promoters on its growth over time. The promoters 'E.coli had been transformed with were PyeaR, M-B and B-M. These are promoters which all react to nitrogenous species. By running these together, we can obtain a direct comparison between all three of these promoters on the growth of E.coli. To see if there are any significant changes, the study was run alongside E.coli cells which had not been transformed with anything. For the rest of this brief report, untransformed cells will be referred to as Alpha cells and the other E.coli cells with transformations will be referred to as the promoter with which they were transformed with.
The E.coli cells used in the study and for the transformation are the same type of cells (Alpha select gold standard cells from Bioline). A colony was inoculated into 5ml of LB media overnight and the cells spun down the following morning and diluted with fresh LB until an OD reading at 600nm of 0.2 ± 0.01 was obtained. Three repeats were made of each sample.
The study lasted for 12 hours. An OD reading at 600nm was taken once an hour. Between the hour, the cuvettes were put into a 37ᵒC incubator to encourage growth and for standardising measurements with other growth studies. To calculate the number of cells in the samples, a calibration curve was set up. This involved using cultures of the E.coli cells without transformations. The E.coli cells were diluted with different volumes of LB and OD readings were taken as well as plating on Agar plates. After a day of growth, the numbers on these plates were counted and recorded. The CFU/ml was calculated. When the OD readings (x axis) and the CFU/ml (y axis) readings are plotted, the equation of the line of best fit, gives a conversion for the absorbance readings. This allowed us to measure the growth. This is demonstrated in figure 1.
Figure 1: Calibration curve to calculate the conversion factor between OD reading at 600nm and the number of colony forming units growing per ml (CFU/ml)
We found that there was a significant difference between Alpha cells and PyeaR cells. Initially, Alpha cells had a greater growth rate, but after the third hour into the study, the growth rate of PyeaR was faster than that of Alpha cells. The overall growth rate of PyeaR cells was significantly faster that Alpha cells (Levenes Test, F = 1.009 p = 0.372; T Test, t = 4.196, df = 4, p = 0.014).
Figure 2: Growth of PyeaR transformed E.coli cells relative to Alpha cell (untransformed cells. Error bars show the standard deviation between the three repeats. For clarity reasons, lines of best fit are not shown
The growth pattern and rate of E.coli cells with or without transformation with B-M and M-B show little difference. Any differences in growth rate were not significant. There was lots of overlap. As previously described, there was a significant difference between the growth rate of PyeaR and Alpha cells. There was also a significant difference between MB/BM and PyeaR cells. The statistical results can be seen in Table 1
Figure 3: Growth over 12 hours of Alpha, M-B and B-M. Error bars and lines of best fit are not shown for clarity reasons.
Table 1: ANOVA readings of statistical differences between Alpha (1) PyeaR (2), MB (3) and BM (4).
From all the above graphs, it can be seen that with the starting concentration of cells as high as they are, the cultures are in exponential stage and do not undergo lag phase. A further growth study will be carried out on purely the lag phase with lower starting concentrations. As the starting absorbances here are approximately 0.2 at a wavelength of 600nm, the lag phase study will involve starting absorbances of 0.04 and lower.
Following the above study, we found that a lag phase only study needed to be carried out to see if there was a significant difference in the lag phase. Again the study protocol was the same except that the starting concentration absorbances at 600nm was lowered to <0.04. It was extremely difficult to keep the absorbances ranges within 0.005 so the range is actually 0.3±0.1. The below graph shows the mean average of all the data; using the data from the calibration curve, the absorbances were converted to colony forming units per ml (CFU/ml). The trend lines of alpha cells, BM/MB and PyeaR transformed cells are shown within this order from highest to lowest trendlines. One single trendline was used to represent BM and MB because the actual trendlines were extremely similar. Using the initial concentrations of 0.3±0.1 showed that there is little difference between the growth rates. Using statistical analysis, it was found that there was no significant difference between any of the transformed cells relative to Alpha cells or to each other (Anova, p > 0.05).
From this study we have found that changes in growth occur during exponential growth phase and not the lag growth phase.
The team plan to carry out a growth study similar to that above for the BM MB, as when working with construct transformed E.coli team members have noticed quick growth . This will allow analysis of any differences in the rate of growth between E.coli transformed with either constructs from the alpha gold control.