Team:Michigan/Parts
From 2012.igem.org
(Difference between revisions)
(Prototype team page) |
|||
| (24 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{:Team:Michigan/Template:New}} | |
<html> | <html> | ||
| - | <div | + | <div class="body"> |
| - | <div id=" | + | <div id="basic"> |
| - | + | <h3> | |
| - | < | + | Parts |
| - | < | + | </h3> |
| - | + | Michigan 2012 #s: K880000-K880005 | |
| - | < | + | <br><br> |
| - | < | + | |
| - | + | <h2>BBa_K880000</html>[http://partsregistry.org/Part:BBa_K880000]<html></h2> | |
| - | < | + | |
| - | < | + | The Hbif recombinase is a putative regulator of the Escherichia coli type 1 fimbriation system, capable of switching the fimS invertible region in the production of fimbriae in a manner similar to the better characterized FimB and FimE recombinases. Orientation control of the fimS switch by recombinases is specific to directionality, with Hbif catalysing the conversion of “OFF” oriented regions to “ON” oriented ones such that the fimS promoter is facing genes responsible for type 1 fimbriae production. |
| + | <br><br> | ||
| + | The hbiF gene was synthesized in vitro as device capable of opposing the directional inversion of the characterized fimE gene K137007 for the purpose of engineering bidirectional molecular switches. A synthetic circuit can be made by fusing two unique invertable repeats (IRs) recognized by the recombinase-parts K137008 and K137010, in that order- flanking a sequence of interest of 200-300bp and expressing hbiF or fimE in low-to-moderate levels to cause the sequence to invert: | ||
| + | <br><br> | ||
| + | -<i>fimE </i>will cause a template strand between the IRs to face the upstream coding strand. | ||
| + | |||
| + | -<i>hbiF</i> will cause a coding strand between the IRs to face the downstream template strand. | ||
| + | <br><br> | ||
| + | Note that IRs’ initial orientation is FimE flippable, but not HbiF flippable. | ||
| + | <br><br> | ||
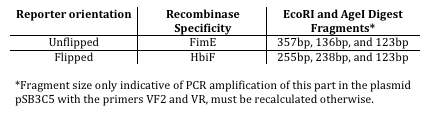
| + | This reaction can be characterized by digesting asymmetrical digest reporter K880001 with EcoRI and AgeI and determining the lengths of the resulting fragments. | ||
| + | <br><br> | ||
| + | Additionally, HbiF contains an RFC25 suffix and is capable of undergoing protein fusions to fluorescent proteins for quantification, affinity purification tags, or degradation tags such as K880004 to increase circuit responsiveness. | ||
| + | <br><br> | ||
| + | Reference: Xie et al. Infect. Immun. 2006, 74(7):4039 | ||
| + | <br><br> | ||
| + | |||
| + | <h2>BBa_K880001</html>[http://partsregistry.org/Part:BBa_K880001]<html></h2> | ||
| + | |||
| + | K880001 is a utility for characterizing the functionality of <i>fim</i> based recombinases such as FimB, FimE, and HbiF and consists of K137008 and K137010 flanking a “junk” DNA sequence K165006 containing an off-center AgeI restriction site proximal to the BioBrick prefix; inversion of the fragment will cause the site to move towards the suffix. | ||
| + | <br><bR> | ||
| + | Digestion of the unflipped reporter with EcoRI and AgeI will yield a ~70bp fragment when in a FimE-flippable orientation and a ~220bp fragment when in an HbiF flippable orientation. Alternatively, digestion with AgeI and PstI will yield fragments of roughly opposite orientation. The reporter in the registry sequence is FimE flippable. | ||
| + | <br><br> | ||
| + | An artificial construct based off the assay performed in Teng et al. Infect. Immun. 2005, 73(5):2923. | ||
| + | |||
</html> | </html> | ||
| + | [[File:MichDiagram1.png|400px|thumb|none]] | ||
| + | <html> | ||
| + | <br><br> | ||
| - | |||
| - | + | <h2>BBa_K880002</html>[http://partsregistry.org/Part:BBa_K880002]<html></h2> | |
| - | + | GFP fluorescent reporter capable of assaying the functionality of fim regulatory recombinases; based on K137058. | |
| - | + | <br><br> | |
| - | + | K137058’s invertible repeat sequences responsible for recombinase binding are ordered incorrectly; the regions are not identical, and have “left” and “right” components. K137008 (invertible repeat-right) and K137010 (invertible repeat-left) are mislabeled and belong in the order “K137008-region to invert-K137010” in accordance with the wild-type fim system. | |
| - | + | <br><br> | |
| - | + | Repeat sequence in this part exist in the correct (wild-type) orientation. | |
| - | + | <br><br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <h2>BBa_K880003</html>[http://partsregistry.org/Part:BBa_K880003]<html></h2> | ||
| + | IRL K137010 + Sequence with AgeI cut-site K165006 + IRR K137008 Mi8 | ||
| - | + | This is an asymmetrically digestible reporter, same orientation of inverted repeats as K137058 (external sites located internally of flip region)(IRL - “flip region” - IRR) for assaying fim-Based recombinase activity. | |
| + | </html> | ||
| - | + | [[File:MichTable1.png|300px|center|thumb]] | |
| + | <html> | ||
| + | </html> | ||
| + | [[File:MichDiagram2.png|400px|thumb|none]] | ||
| + | <html> | ||
| + | <br> | ||
| + | <h2>BBa_K880004</html>[http://partsregistry.org/Part:BBa_K880004]<html></h2> | ||
| + | -LVA Tag in RFC25 | ||
| + | <br><br> | ||
| + | The amino acid sequence AANDENYALVA can be fused onto proteins in order to rapidly reduce their half life in the cytoplasm. Responsiveness for genetic circuits can be improved by c-terminal fusions of this short, handy sequence. | ||
| + | <br><bR> | ||
| + | This part possesses an RFC25 prefix to facilitate in-frame translation. | ||
| + | <br><br> | ||
| + | Reference: Andersen et al. Appl Environ Microbiol. 1998 June; 64(6): 2240–2246. | ||
| + | <br><br> | ||
| - | <groupparts>iGEM012 Michigan</groupparts> | + | <h2>BBa_K880005</html>[http://partsregistry.org/Part:BBa_K880005]<html></h2> |
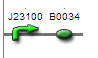
| + | -J23100+B0034 | ||
| + | -Strong promoter, strong RBS combination for high expression levels of proteins | ||
| + | <br><br> | ||
| + | |||
| + | Consensus constitutive promoter and RBS sequence-produces strongest possible expression of given downstream genes. | ||
| + | <br><br> | ||
| + | </html> | ||
| + | [[File:MichParts0005.png|400px|thumb|center]] | ||
| + | <html> | ||
| + | |||
| + | <h2>Parts Reviewed</h2> | ||
| + | -BBa_K137058</html>[http://partsregistry.org/Part:BBa_K137058:Experience#User_Reviews]<html><br> | ||
| + | -BBa_K137007</html>[http://partsregistry.org/Part:BBa_K137007:Experience#User_Reviews]<html><br> | ||
| + | -pSB4c5</html>[http://partsregistry.org/Part:pSB4C5:Experience#User_Reviews]<html><br> | ||
| + | -BBa_B0015</html>[http://partsregistry.org/Part:BBa_B0015:Experience#User_Reviews]<html><br> | ||
| + | -BBa_K137080</html>[http://partsregistry.org/Part:K137080:Experience#User_Reviews]<html><br> | ||
| + | -BBa_K113009</html>[http://partsregistry.org/Part:BBa_K113009:Experience#User_Reviews]<html><br> | ||
| + | -BBa_K206000</html>[http://partsregistry.org/Part:BBa_K206000:Experience#User_Reviews]<html><br> | ||
| + | -BBa_K206001</html>[http://partsregistry.org/Part:BBa_K206001:Experience#User_Reviews]<html><br> | ||
| + | -BBa_I13453</html>[http://partsregistry.org/Part:BBa_I13453:Experience#User_Reviews]<html><br> | ||
| + | |||
| + | <br><br> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | </html> | ||
| + | <center> | ||
| + | <groupparts>iGEM012 Michigan</groupparts></center> | ||
Latest revision as of 02:54, 4 October 2012
Parts
Michigan 2012 #s: K880000-K880005BBa_K880000[1]
The Hbif recombinase is a putative regulator of the Escherichia coli type 1 fimbriation system, capable of switching the fimS invertible region in the production of fimbriae in a manner similar to the better characterized FimB and FimE recombinases. Orientation control of the fimS switch by recombinases is specific to directionality, with Hbif catalysing the conversion of “OFF” oriented regions to “ON” oriented ones such that the fimS promoter is facing genes responsible for type 1 fimbriae production.The hbiF gene was synthesized in vitro as device capable of opposing the directional inversion of the characterized fimE gene K137007 for the purpose of engineering bidirectional molecular switches. A synthetic circuit can be made by fusing two unique invertable repeats (IRs) recognized by the recombinase-parts K137008 and K137010, in that order- flanking a sequence of interest of 200-300bp and expressing hbiF or fimE in low-to-moderate levels to cause the sequence to invert:
-fimE will cause a template strand between the IRs to face the upstream coding strand. -hbiF will cause a coding strand between the IRs to face the downstream template strand.
Note that IRs’ initial orientation is FimE flippable, but not HbiF flippable.
This reaction can be characterized by digesting asymmetrical digest reporter K880001 with EcoRI and AgeI and determining the lengths of the resulting fragments.
Additionally, HbiF contains an RFC25 suffix and is capable of undergoing protein fusions to fluorescent proteins for quantification, affinity purification tags, or degradation tags such as K880004 to increase circuit responsiveness.
Reference: Xie et al. Infect. Immun. 2006, 74(7):4039
BBa_K880001[2]
K880001 is a utility for characterizing the functionality of fim based recombinases such as FimB, FimE, and HbiF and consists of K137008 and K137010 flanking a “junk” DNA sequence K165006 containing an off-center AgeI restriction site proximal to the BioBrick prefix; inversion of the fragment will cause the site to move towards the suffix.Digestion of the unflipped reporter with EcoRI and AgeI will yield a ~70bp fragment when in a FimE-flippable orientation and a ~220bp fragment when in an HbiF flippable orientation. Alternatively, digestion with AgeI and PstI will yield fragments of roughly opposite orientation. The reporter in the registry sequence is FimE flippable.
An artificial construct based off the assay performed in Teng et al. Infect. Immun. 2005, 73(5):2923.
BBa_K880002[3]
GFP fluorescent reporter capable of assaying the functionality of fim regulatory recombinases; based on K137058.K137058’s invertible repeat sequences responsible for recombinase binding are ordered incorrectly; the regions are not identical, and have “left” and “right” components. K137008 (invertible repeat-right) and K137010 (invertible repeat-left) are mislabeled and belong in the order “K137008-region to invert-K137010” in accordance with the wild-type fim system.
Repeat sequence in this part exist in the correct (wild-type) orientation.
BBa_K880003[4]
IRL K137010 + Sequence with AgeI cut-site K165006 + IRR K137008 Mi8 This is an asymmetrically digestible reporter, same orientation of inverted repeats as K137058 (external sites located internally of flip region)(IRL - “flip region” - IRR) for assaying fim-Based recombinase activity.
BBa_K880004[5]
-LVA Tag in RFC25The amino acid sequence AANDENYALVA can be fused onto proteins in order to rapidly reduce their half life in the cytoplasm. Responsiveness for genetic circuits can be improved by c-terminal fusions of this short, handy sequence.
This part possesses an RFC25 prefix to facilitate in-frame translation.
Reference: Andersen et al. Appl Environ Microbiol. 1998 June; 64(6): 2240–2246.
BBa_K880005[6]
-J23100+B0034 -Strong promoter, strong RBS combination for high expression levels of proteinsConsensus constitutive promoter and RBS sequence-produces strongest possible expression of given downstream genes.
Parts Reviewed
-BBa_K137058[7]-BBa_K137007[8]
-pSB4c5[9]
-BBa_B0015[10]
-BBa_K137080[11]
-BBa_K113009[12]
-BBa_K206000[13]
-BBa_K206001[14]
-BBa_I13453[15]
 "
"