Team:Michigan/Modeling
From 2012.igem.org
Modeling
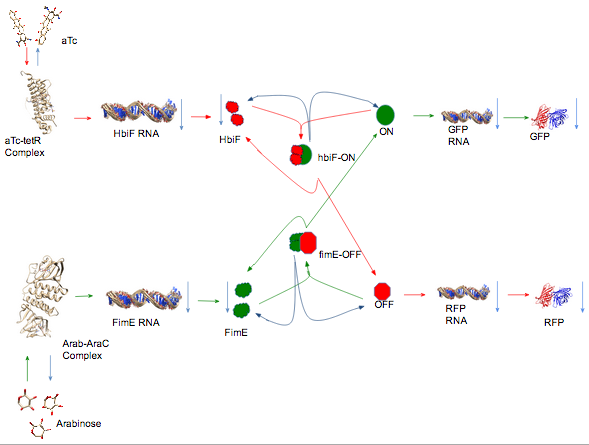
Modeling is a way of figuring out how your system works and how it produces the behavior that it does. It is also a way of predicting the outcome of your system. Models are never completely realistic. They are approximations of your system and are only as accurate as the your assumptions. You approximate your system by making assumptions that you either recognize to be true or are complexities that you assume to be unimportant. A good model can be useful in representing the behavior of your system.We chose to model our system using a Mass Action model that utilizes a system of Ordinary Differential Equations (ODE).
In all of our equations, we assume that the cell always has a constant amount of NTP’s, amino acids, ribosomes, and other basic necessities. Since we are assuming these to be constant, their effects on amounts of RNA, DNA, protein, etc... are abstracted away into the rate constants.
We start by considering the amount of inducer present in a bound and unbound state.
$\frac{d[Arab*AraC]}{dt} = ( f_{A} * (Arab_{tot} - [Arab*AraC]) ) - (di_{A}*[Arab*AraC])$
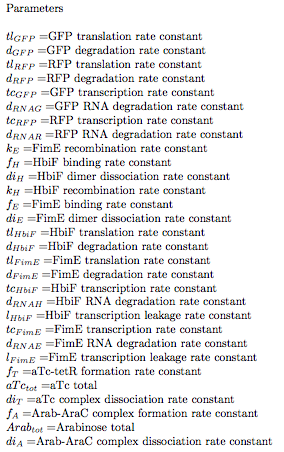
$[Arab*AraC] =$ Concentration of Arabinose bound to the AraC repressor protein (Arab-AraC Complex)
$f_{A} = $ Arab-AraC complex formation rate constant
$Arab_{tot} = $ Arabinose total
$di_{A} = $ Arab-AraC complex dissociation rate constant
We will assume that the total amount of inducer and repressor is constant. We will also assume that there is always more repressor then there is arabinose to bind to the repressor. Thus, the amount of Arabinose present is $Arab_{tot} - [Arab*AraC]$ and the rate of repressor complex production is proportional to the amount of Arabinose available: $f_{A} * (Arab_{tot} - [Arab*AraC]$. The rate of repressor complex dissociation is proportional to the amount of Arab-AraC complex available: $di_{A}*[Arab*AraC]$. The rate of change of Arab-AraC complex is simply the rate of formation minus the rate of dissociation. The equation for the change in aTc-tetR complex was formulated in the same manner.
Next, we consider the amount of recombinase RNA produced.
$\frac{d[RNA_{FimE}]}{dt} = ( (tc_{FimE}*[Arab*AraC]) - (d_{RNAE}*[RNA_{FimE}]) + (l_{FimE}) )$
$[RNA_{FimE}] =$ concentration of fimE RNA
$tc_{FimE} = $ FimE transcription rate constant
$d_{RNAE} = $ FimE RNA degradation rate constant
$l_{FimE} = $ FimE transcription leakage rate constant
We consider the rate of formation of fimE RNA to be proportional to the concentration of Arab-AraC complex, $tc_{FimE}*[Arab*AraC]$. Higher $[Arab*AraC]$ will result in more unbound repressor and therefore more transcription of fimE. The strength of the inducible promoter and the RBS for the recombinase will affect the transcription rate; however, we chose to abstract away these details into the fimE transcription rate constant $tc_{FimE}$. DEgradation of RNA will also affect the concentration of fimE RNA. We consider degradation to be proportional to the concentration of fimE RNA, $d_{RNAE}*[RNA_{FimE}]$. Finally, since all inducible promoter systems have basal expression, we add a leakage transcription rate $l_{FimE}$ that is specific to the promoter, RBS, and the gene being transcribed. The rate of change of fimE RNA present is the rate of RNA formation minus the rate of RNA degradation plus the rate of leaky transcription. The equation for the change in hbiF RNA was derived in the same manner.
Once we have considered RNA, we must consider the amount of protein. Continuing with fimE as our model recombinase, we have:
$\frac{d[FimE]}{dt} = (tl_{FimE}*[RNA_{FimE}]) - (d_{FimE}*[FimE]) - (f_{E}*[OFF]*([FimE]^2)) + (k_{E}*[OFF*FimE^2]) + (di_{E}*[OFF*FimE^2])$
$[FimE] =$ concentration of fimE protein available for binding to DNA
$[FimE]^2 =$ concentration of fimE dimers, the exponent represents that 2 fimE proteins are required to make functioning recombinase unit
$[OFF * FimE^2] =$ represents the concentration of fimE recombinase bound to OFF state DNA
$[OFF] =$ the concentration of DNA in the “OFF state”, DNA that is invertible by fimE
$tl_{FimE} = $FimE translation rate constant
$d_{FimE} = $FimE degradation rate constant
$f_{E} = $FimE binding rate constant
$k_{E} = $FimE recombination rate constant
$di_{E} = $FimE dimer dissociation rate constant
Based on previous knowledge of how recombinases function, we assumed that fimE needs to form a dimer in order to be active $[FimE]^2$. Rather than keeping track of how much fimE was in a dimer state, we decided to account for both dimers and monomers as simply fimE protein. The first and second terms in the equation are straightforward translation and degradation rates of fimE protein. As more fimE dimers form and bind to OFF state DNA, the amount of fimE protein available for binding decreases, $f_{E}*[OFF]*([FimE]^2)$. The $[FimE]$ increases when either the bound fimE flips the OFF state DNA to the ON state, or the fimE dissociates from the OFF state DNA without flipping: $(k_{E}*[OFF*FimE^2]) + (di_{E}*[OFF*FimE^2])$. Again, the equation for the change in hbiF protein over time is a mirror image of this equation.
Next, we write the equation for the amount of fimE bound to OFF state DNA.
$\frac{d[OFF*FimE^2]}{dt} = (f_{E}*[OFF]*([FimE]^2)) - (k_{E}*[OFF*FimE^2]) - (di_{E}*[OFF*FimE^2])$
This is simply a trivial matter of changing the signs of the last three terms in the fimE protein equation.
Next, we consider the amount of DNA in each state that is available to be bound by a recombinase. We consider only the OFF state DNA here. The ON state equation is a mirror image.
$\frac{d[OFF]}{dt} = (k_{H}*[ON*HbiF^2]) - (f_{E}*[OFF]*([FimE]^2)) + (di_{E}*[OFF*FimE^2])$
$[ON* HbiF^2] =$ represents the concentration of hbiF recombinase bound to OFF state DNA
$k_{H} = $ HbiF recombination rate constant
We are considering HbiF flippable DNA to be ON state DNA. When hbiF dimers bind to ON state DNA and flip the DNA, the ON state DNA becomes OFF state DNA: $k_{H}*[ON*HbiF^2]$. Since we $[OFF]$ only represents unbound OFF state DNA, the binding of fimE dimers to the DNA decreases $[OFF]$: $f_{E}*[OFF]*([FimE]^2)$. Finally, dissociation of fimE dimers bound to OFF state DNA results in unbound OFF state DNA: $di_{E}*[OFF*FimE^2]$.
Now that we have worked our way from induction of recombinase production to flipping of the DNA, we consider the production of protein by the promoter between the recombinase sites.
$\frac{d[RNA_{GFP}]}{dt} = (tc_{GFP}*[ON]) - (d_{RNAG}*[RNA_{GFP}])$
$\frac{d[GFP]}{dt} = (tl_{GFP}*[RNA_{GFP}]) - (d_{GFP}*[GFP])$
$[RNA_{GFP}] =$ concentration of GFP RNA
$[GFP] =$ concentration of GFP
$[ON] =$ the concentration of DNA in the “ON state”, DNA that is invertible by hbiF
$tc_{GFP} = $GFP transcription rate constant
$d_{RNAG} = $GFP RNA degradation rate constant
$tl_{GFP} = $GFP translation rate constant
$d_{GFP} = $GFP degradation rate constant
The rate of GFP RNA production is proportional to the concentration of ON state DNA: $tc_{GFP}*[ON]$. The strength of the promoter and RBS expressing GFP is abstracted away into the GFP transcription rate constant $tc_{GFP}$. The rate of degradation of GFP RNA is proportional to the concentration of GFP RNA: $d_{RNAG}*[RNA_{GFP}]$. The degradation rate of GFP protein is treated in a similar manner. The rate of GFP production is proportional to the concentration of GFP RNA: $tl_{GFP}*[RNA_{GFP}]$. The equations for RFP production are mirror images of these two equations.
The equations are listed below:
We start by considering the amount of inducer present in a bound and unbound state.
$\frac{d[Arab*AraC]}{dt} = ( f_{A} * (Arab_{tot} - [Arab*AraC]) ) - (di_{A}*[Arab*AraC])$
$[Arab*AraC] =$ Concentration of Arabinose bound to the AraC repressor protein (Arab-AraC Complex)
$f_{A} = $ Arab-AraC complex formation rate constant
$Arab_{tot} = $ Arabinose total
$di_{A} = $ Arab-AraC complex dissociation rate constant
We will assume that the total amount of inducer and repressor is constant. We will also assume that there is always more repressor then there is arabinose to bind to the repressor. Thus, the amount of Arabinose present is $Arab_{tot} - [Arab*AraC]$ and the rate of repressor complex production is proportional to the amount of Arabinose available: $f_{A} * (Arab_{tot} - [Arab*AraC]$. The rate of repressor complex dissociation is proportional to the amount of Arab-AraC complex available: $di_{A}*[Arab*AraC]$. The rate of change of Arab-AraC complex is simply the rate of formation minus the rate of dissociation. The equation for the change in aTc-tetR complex was formulated in the same manner.
Next, we consider the amount of recombinase RNA produced.
$\frac{d[RNA_{FimE}]}{dt} = ( (tc_{FimE}*[Arab*AraC]) - (d_{RNAE}*[RNA_{FimE}]) + (l_{FimE}) )$
$[RNA_{FimE}] =$ concentration of fimE RNA
$tc_{FimE} = $ FimE transcription rate constant
$d_{RNAE} = $ FimE RNA degradation rate constant
$l_{FimE} = $ FimE transcription leakage rate constant
We consider the rate of formation of fimE RNA to be proportional to the concentration of Arab-AraC complex, $tc_{FimE}*[Arab*AraC]$. Higher $[Arab*AraC]$ will result in more unbound repressor and therefore more transcription of fimE. The strength of the inducible promoter and the RBS for the recombinase will affect the transcription rate; however, we chose to abstract away these details into the fimE transcription rate constant $tc_{FimE}$. DEgradation of RNA will also affect the concentration of fimE RNA. We consider degradation to be proportional to the concentration of fimE RNA, $d_{RNAE}*[RNA_{FimE}]$. Finally, since all inducible promoter systems have basal expression, we add a leakage transcription rate $l_{FimE}$ that is specific to the promoter, RBS, and the gene being transcribed. The rate of change of fimE RNA present is the rate of RNA formation minus the rate of RNA degradation plus the rate of leaky transcription. The equation for the change in hbiF RNA was derived in the same manner.
Once we have considered RNA, we must consider the amount of protein. Continuing with fimE as our model recombinase, we have:
$\frac{d[FimE]}{dt} = (tl_{FimE}*[RNA_{FimE}]) - (d_{FimE}*[FimE]) - (f_{E}*[OFF]*([FimE]^2)) + (k_{E}*[OFF*FimE^2]) + (di_{E}*[OFF*FimE^2])$
$[FimE] =$ concentration of fimE protein available for binding to DNA
$[FimE]^2 =$ concentration of fimE dimers, the exponent represents that 2 fimE proteins are required to make functioning recombinase unit
$[OFF * FimE^2] =$ represents the concentration of fimE recombinase bound to OFF state DNA
$[OFF] =$ the concentration of DNA in the “OFF state”, DNA that is invertible by fimE
$tl_{FimE} = $FimE translation rate constant
$d_{FimE} = $FimE degradation rate constant
$f_{E} = $FimE binding rate constant
$k_{E} = $FimE recombination rate constant
$di_{E} = $FimE dimer dissociation rate constant
Based on previous knowledge of how recombinases function, we assumed that fimE needs to form a dimer in order to be active $[FimE]^2$. Rather than keeping track of how much fimE was in a dimer state, we decided to account for both dimers and monomers as simply fimE protein. The first and second terms in the equation are straightforward translation and degradation rates of fimE protein. As more fimE dimers form and bind to OFF state DNA, the amount of fimE protein available for binding decreases, $f_{E}*[OFF]*([FimE]^2)$. The $[FimE]$ increases when either the bound fimE flips the OFF state DNA to the ON state, or the fimE dissociates from the OFF state DNA without flipping: $(k_{E}*[OFF*FimE^2]) + (di_{E}*[OFF*FimE^2])$. Again, the equation for the change in hbiF protein over time is a mirror image of this equation.
Next, we write the equation for the amount of fimE bound to OFF state DNA.
$\frac{d[OFF*FimE^2]}{dt} = (f_{E}*[OFF]*([FimE]^2)) - (k_{E}*[OFF*FimE^2]) - (di_{E}*[OFF*FimE^2])$
This is simply a trivial matter of changing the signs of the last three terms in the fimE protein equation.
Next, we consider the amount of DNA in each state that is available to be bound by a recombinase. We consider only the OFF state DNA here. The ON state equation is a mirror image.
$\frac{d[OFF]}{dt} = (k_{H}*[ON*HbiF^2]) - (f_{E}*[OFF]*([FimE]^2)) + (di_{E}*[OFF*FimE^2])$
$[ON* HbiF^2] =$ represents the concentration of hbiF recombinase bound to OFF state DNA
$k_{H} = $ HbiF recombination rate constant
We are considering HbiF flippable DNA to be ON state DNA. When hbiF dimers bind to ON state DNA and flip the DNA, the ON state DNA becomes OFF state DNA: $k_{H}*[ON*HbiF^2]$. Since we $[OFF]$ only represents unbound OFF state DNA, the binding of fimE dimers to the DNA decreases $[OFF]$: $f_{E}*[OFF]*([FimE]^2)$. Finally, dissociation of fimE dimers bound to OFF state DNA results in unbound OFF state DNA: $di_{E}*[OFF*FimE^2]$.
Now that we have worked our way from induction of recombinase production to flipping of the DNA, we consider the production of protein by the promoter between the recombinase sites.
$\frac{d[RNA_{GFP}]}{dt} = (tc_{GFP}*[ON]) - (d_{RNAG}*[RNA_{GFP}])$
$\frac{d[GFP]}{dt} = (tl_{GFP}*[RNA_{GFP}]) - (d_{GFP}*[GFP])$
$[RNA_{GFP}] =$ concentration of GFP RNA
$[GFP] =$ concentration of GFP
$[ON] =$ the concentration of DNA in the “ON state”, DNA that is invertible by hbiF
$tc_{GFP} = $GFP transcription rate constant
$d_{RNAG} = $GFP RNA degradation rate constant
$tl_{GFP} = $GFP translation rate constant
$d_{GFP} = $GFP degradation rate constant
The rate of GFP RNA production is proportional to the concentration of ON state DNA: $tc_{GFP}*[ON]$. The strength of the promoter and RBS expressing GFP is abstracted away into the GFP transcription rate constant $tc_{GFP}$. The rate of degradation of GFP RNA is proportional to the concentration of GFP RNA: $d_{RNAG}*[RNA_{GFP}]$. The degradation rate of GFP protein is treated in a similar manner. The rate of GFP production is proportional to the concentration of GFP RNA: $tl_{GFP}*[RNA_{GFP}]$. The equations for RFP production are mirror images of these two equations.
The equations are listed below:
 "
"