Team:Macquarie Australia/Results
From 2012.igem.org
| (87 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<html> | <html> | ||
<h1><center>Results and Characterisation</h1></center> | <h1><center>Results and Characterisation</h1></center> | ||
| - | + | <body> | |
<p><center>To quickly skip to the section that you wish to read, click on the links below.</p></center> | <p><center>To quickly skip to the section that you wish to read, click on the links below.</p></center> | ||
<center><table> | <center><table> | ||
<th colspan="2"> | <th colspan="2"> | ||
| - | <center><font color=#c85a17 size="5"><b>Results</b></font></center> | + | <center><font color=#c85a17 size="5"><b>Results</b></font></center></th> |
| - | </th> | + | <tr><td><a href="#1"><h3>Heme Oxygenase</h3></a></td> |
| - | + | <td><a href=#3><h3>Bacteriophytochromes</h3></a> | |
| - | <td> | + | |
| - | <a href="#1"><h3>Heme Oxygenase</h3></a> | + | |
| - | </td> | + | |
| - | <td> | + | |
| - | <a href=#3><h3>Bacteriophytochromes</h3></a> | + | |
</td></tr></table><br> | </td></tr></table><br> | ||
| - | <table><th colspan="2"><center><font color=#c85a17 size="5">Sequencing Results</font | + | <table><th colspan="2"><center><font color=#c85a17 size="5">Sequencing Results</font></th> |
| - | <tr> | + | <tr><td><a href="#5"><h3>Heme Oxygenase</h3></a></td> |
| - | <td> | + | <td><a href="#6"><h3>Bacteriophytochromes</h3></a></td></tr> |
| - | <a href="#5"><h3>Heme Oxygenase</h3></a> | + | |
| - | </td> | + | |
| - | <td> | + | |
| - | <a href="#6"><h3>Bacteriophytochromes</h3></a | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</table> | </table> | ||
| + | <br> | ||
| + | <table><center><th colspan="4"><font color=#c85a17 size="5">Characterisation</font></th></center> | ||
| + | <tr><td><a href="#2"><h3>Heme Oxygenase</h3></a></td> | ||
| + | <td><a href="#4"><h3>Bacteriophytochromes</h3></a></td> | ||
| + | <td><a href="#7"><h3>Building The Switch</h3></td> | ||
| + | <td><a href="#switch"><h3>Reversing The Switch</h3></td></tr> | ||
| + | </table></center> | ||
<hr> | <hr> | ||
| - | <center><a name="1"><h3>Heme Oxygenase Results</h3></center> | + | <center><a name="1"><h3>Heme Oxygenase Results</h3></a></center> |
<p>We produced a Heme Oxygenase BioBrick that was codon optimized for <i>E. coli</i>. The Gibson assembly of the T7 promoter containing Heme Oxygenase was successful. The transformation was successful with numerous colonies grown using Chloramphenicol as the selecting agent. Six colonies were selected and then they were sequenced before digestion with EcoR1 and Spe1. The sequencing showed that all of the colonies contained the plasmid with a Heme oxygenase identical to the original protein sequence. The gel containing the digested Heme Oxygenase bearing plasmid can be seen in Figure 1.</p><br> | <p>We produced a Heme Oxygenase BioBrick that was codon optimized for <i>E. coli</i>. The Gibson assembly of the T7 promoter containing Heme Oxygenase was successful. The transformation was successful with numerous colonies grown using Chloramphenicol as the selecting agent. Six colonies were selected and then they were sequenced before digestion with EcoR1 and Spe1. The sequencing showed that all of the colonies contained the plasmid with a Heme oxygenase identical to the original protein sequence. The gel containing the digested Heme Oxygenase bearing plasmid can be seen in Figure 1.</p><br> | ||
<center><table><tr><td> | <center><table><tr><td> | ||
| Line 41: | Line 28: | ||
</td></tr> | </td></tr> | ||
<tr><td> | <tr><td> | ||
| - | Figure 1: The restriction digest showing the linearised plasmid backbone (Black Box)<br> | + | <center>Figure 1: The restriction digest showing the linearised plasmid backbone (Black Box)<br> |
| - | and the heme oxygenese gene (Green Box). We used a 1kb ladder. | + | and the heme oxygenese gene (Green Box). We used a 1kb ladder |
| + | <br>(in increasing Kbp: 0.25, 0.5, 0.75, 1, 1.5, 2, 2.5, 3, 4, 5, 6, 8, 10).</center> | ||
</td></tr></table></center> | </td></tr></table></center> | ||
| - | <p>The sequencing results for the six plasmids above can be seen below.</p> | + | <p>The lower band is the T7 Heme Oxygenase BioBrick (771 bp) which was expected. Similarly, the plasmid backbone was visible at the expected length (2070 bp). The similarity in intensity of the bands also demonstrates that the assembly and the digest were successful. The sequencing results for the six plasmids above can be seen below.</p> |
<hr> | <hr> | ||
| - | <center><a name="5"><h3>Heme Oxygenase Sequencing Results</h3></center> | + | <center><a name="5"><h3>Heme Oxygenase Sequencing Results</h3></a></center> |
| - | <p>The | + | <p>The Gibson Assembly performed was transformed into 6 different cultures of competent cells. Three of theses were selected for sequencing using both forward and reverse primers. They were sequenced using the forward and reverse primers for the BioBricks. We performed Blastx and Blastn pipelines (available <a href="http://blast.ncbi.nlm.nih.gov/">here</a>) to determine if there was a significant change in the protein sequence and to determine the identity of the plasmid. The sequences below are named to show: that it is heme oxygenase (1C), which transformation (4,5,6), and whether it is the forward or reverse primer (F or R).</p><br> |
<center><table border="3" cellpadding="4" cellspacing="0"> | <center><table border="3" cellpadding="4" cellspacing="0"> | ||
| - | <tr><td>Sample</td><td> | + | <tr><td>Sample</td><td>Identity</td> <td>E-value</td><td>MaxID</td></tr> |
<tr><td>1C-6F</td><td>Heme Oxygenase (Synechocystic sp. PCC603)</td><td>7e-176</td><td>99%</td></tr> | <tr><td>1C-6F</td><td>Heme Oxygenase (Synechocystic sp. PCC603)</td><td>7e-176</td><td>99%</td></tr> | ||
<tr><td>1C-6R</td><td>Heme Oxygenase (Synechocystic sp. PCC603)</td><td>1e-171</td><td>99%</td></tr> | <tr><td>1C-6R</td><td>Heme Oxygenase (Synechocystic sp. PCC603)</td><td>1e-171</td><td>99%</td></tr> | ||
<tr><td>1C-4F</td><td>Heme Oxygenase (Synechocystic sp. PCC603)</td><td>4e-176</td><td>99%</td></tr> | <tr><td>1C-4F</td><td>Heme Oxygenase (Synechocystic sp. PCC603)</td><td>4e-176</td><td>99%</td></tr> | ||
<tr><td>1C-4R</td><td>Heme Oxygenase (Synechocystic sp. PCC603)</td><td>6e-172</td><td>99%</td></tr> | <tr><td>1C-4R</td><td>Heme Oxygenase (Synechocystic sp. PCC603)</td><td>6e-172</td><td>99%</td></tr> | ||
| - | <tr><td>1C-5F</td><td>Heme Oxygenase (Synechocystic sp. PCC603)</td><td> | + | <tr><td>1C-5F</td><td>Heme Oxygenase (Synechocystic sp. PCC603)</td><td>6e-172</td><td>99%</td></tr></table></center><br> |
| - | <tr><td> | + | <center><table><tr><td><ul><li>We successfully assembled the codon optimised Heme Oxygenase.</li> |
| - | <p> | + | <li>The probability that it is not Heme Oxygenase is negligibly small.</li> |
| + | <li>The MaxID score is not 100% due to sequence misreads producing small gaps in the sequence.</li> | ||
| + | <li>The E-value signifies the possibility of matches based purely on chance. A score of | ||
| + | <br>0 identifies no background noise and is expected to be an error-free match.</li> | ||
| + | </ul></td></tr></table></center> | ||
| + | <p>The source of our gene was identified in the sequencing result which showed that the sequencing was accurate. We then compared to the original gBlock sequence and determined that the sequencing was accurate and confirmed the identity of the plasmid. The Blastn pipeline indicated that there was no significant change from the theoretical sequence. With this data we would assume that the protein would be functional and performed assays to determine if this was the case.</p> | ||
<hr> | <hr> | ||
| - | <center><a name="2"><h3>Characterisation of Heme Oxygenase</h3></center> | + | <center><a name="2"><h3>Characterisation of Heme Oxygenase</h3></a></center> |
| - | <p>The T7 | + | <p>The T7 regulated Heme Oxygenase produced was characterised to determine its functionality. BL21 <i>E. coli</i> was transformed with the plasmid, selected for using chloramphenicol, and a culture was inoculated. The culture was then induced with ALA (d-aminolevulinic acid) for the heme pathway and IPTG to promote protein production. They were incubated overnight and the cells were spun down. We observed a functional Heme Oxygenase with the cells appeared a vibrant green after induction by ALA and IPTG. We observed this as well in our assembled switch. The image below demonstrates the green pigment produced by the cells compared to uninduced Heme Oxygenase and the bacteriophytochrome.</p> |
| - | < | + | <center><img src="http://25.media.tumblr.com/tumblr_mazgigGnrs1rg4kjpo1_500.jpg" width=80%></center> |
| + | <br> | ||
| + | <p>The cells were lysed and the protein extract run on an SDS page gel. The gel showed the protein bands near the expected size (27 kDa). This, along with our pigmentation, showed that the BioBrick was producing the functional protein. | ||
| + | <center><img src="https://static.igem.org/mediawiki/2012/7/7b/Cbms-teaching_2012-09-24-HO.jpeg" width=80%></center> | ||
<hr> | <hr> | ||
| - | + | <center><a name="3"><h3>Bacteriophytochromes Results</h3></a></center> | |
| - | <center><a name="3"><h3>Bacteriophytochromes Results</h3></center> | + | <p>Like the Heme Oxygenase, the bacteriophytochromes from <i>Deinococcus radiodurans</i> and <i>Agrobacterium tumefaciens</i> were codon optimised for use in <i>E. Coli</i>. We did not use genomic DNA or previous Macquarie University team DNA at any stage, everything was performed with synthetic DNA we designed. The identity of the plasmid was determined by sequencing and by digestion. The bacteriophytochromes were characterised by demonstrating their ability to bind biliverdin and by their characteristic absorption profile.</p> |
| - | <p>Like the Heme Oxygenase, the bacteriophytochromes from <i>Deinococcus radiodurans</i> and <i>Agrobacterium tumefaciens</i> were codon optimised for use in <i>E. Coli</i>. The identity of the plasmid was determined by sequencing and by digestion.</p> | + | |
<center><img src="https://static.igem.org/mediawiki/2012/7/79/Gel22209.jpg" width=600 height=400></center> | <center><img src="https://static.igem.org/mediawiki/2012/7/79/Gel22209.jpg" width=600 height=400></center> | ||
| - | <p>The digest above shows only | + | <center><table><tr><td>The restriction enzymes used in this digestion were,</tr></td> |
| - | + | <tr><td><ul><li>X= Xba1 P= Pst</li><li>E= EcoR1 S=Spe1</ul></td></tr> | |
| + | <tr><td>We used a 1kb ladder (in increasing Kbp: 0.25, 0.5, 0.75, 1, 1.5, 2, 2.5, 3, 4, 5, 6, 8, 10).</tr></td></table></center> | ||
| + | <p>The digest above shows only shows one band for the different digests performed on the Agro T7 plasmid. The length of the vector and the bacteriophytochrome are similar and make it difficult to resolve the two bands. Only the digested plasmid can be seen which indicates that the bacteriophytochrome component was assembled correctly. Based on the length in base pairs of the fragments (2.25 kbp) we determined that assembly was successful. The first Agro with no T7 digest (asterixed band) shows incomplete digestion of a BioBrick plasmid. This enacted as a control and indicated the size of the plasmid.</p> | ||
<hr> | <hr> | ||
| - | <center><a name="6"><h3>Bacteriophytochrome Sequencing</h3></center> | + | <center><a name="6"><h3>Bacteriophytochrome Sequencing</h3></a></center> |
| - | <p></p> | + | <p>Plasmids extracted from the different transformations were sequenced. </p> |
| + | |||
<center><table border="3" cellpadding="4" cellspacing="0"> | <center><table border="3" cellpadding="4" cellspacing="0"> | ||
| - | <tr><td>Sequence</td><td> | + | <tr><td>Sequence</td><td>Expected Identity</td><td>Identity</td><td>E value</td><td>Max ID</td></tr> |
| - | <tr | + | <tr><td>F3CE1</td><td>Deinococcus radiodurans Bacteriophytochrome</td><td>Chain A, Crystal Structure Of A Monomeric Infrared Fluorescent <br>Deinococcus Radiodurans Bacteriophytochrome Chromophore Binding Domain</td><td>3.00E-160</td><td>99%</td></tr> |
| - | <tr | + | <tr><td>F3CE2</td><td>Deinococcus radiodurans Bacteriophytochrome</td><td>Chain A, Crystal Structure Of A Monomeric Infrared Fluorescent <br>Deinococcus Radiodurans Bacteriophytochrome Chromophore Binding Domain</td><td>2.00E-161</td><td>99%</tr></td> |
| - | + | <tr><td>F3CE3</td><td>Deinococcus radiodurans Bacteriophytochrome</td><td>Chain A, Crystal Structure Of A Monomeric Infrared Fluorescent <br>Deinococcus Radiodurans Bacteriophytochrome Chromophore Binding Domain</td><td>8.00E-163</td><td>99%</tr></td> | |
| - | <tr><td>F3CE3</td><td>Chain A, Crystal Structure Of A Monomeric Infrared Fluorescent <br>Deinococcus Radiodurans Bacteriophytochrome Chromophore Binding Domain</td><td>8.00E-163</td><td>99%</tr></td> | + | <tr><td>R4KE2</td><td>Agrobacterium tumefaciens Bacteriophytochrome</td><td>bacteriophytochrome protein [Agrobacterium tumefaciens str. C58]</td><td>0</td><td>99%</td></tr> |
| - | <tr | + | <tr><td>R5CE1</td><td>Agrobacterium tumefaciens Bacteriophytochrome</td><td>bacteriophytochrome protein [Agrobacterium tumefaciens str. C58]</td><td>0</td><td>99%</td></tr> |
| - | + | <tr><td>R5CE3</td><td>Agrobacterium tumefaciens Bacteriophytochrome</td><td>bacteriophytochrome protein [Agrobacterium tumefaciens str. C58]</td><td>0</td><td>99%</td></tr></table></center><br> | |
| - | <tr><td> | + | |
| - | + | <center><table><tr><td><ul> | |
| - | + | <li>We successfully produced the bacteriophytochromes with protein sequences matching the theoretical sequence.</li><li>The difference in identity is due to the codon optimisation performed. It was also affected by short sequence reads.</li> | |
| - | <tr><td> | + | <li>The 0 E value shows that we have produced the appropriate bacteriophytochrome.</li> |
| - | + | </ul></td></tr></table></center> | |
| - | + | <p>The The Blastx pipeline showed that there was an identical match to the orginal source. This shows that the Gibson Assembly reactions were successful and we expect the protein to be functional. Blastn were run to determine the deviance from the theoretical sequence. The Blastn searches produced indicated that the sequences were nearly identical. The changed bases were examined on the sequencing output and determined to be possible misreads.</p> | |
| - | < | + | |
| - | <li>The difference in identity is due to the codon optimisation performed. It was also affected by short | + | |
| - | <p>The The Blastx pipeline showed that there was | + | |
<hr> | <hr> | ||
| - | <center><a name="4"><h3>Bacteriophytochrome Characterisation</h3></center> | + | <center><a name="4"><h3>Bacteriophytochrome Characterisation</h3></a></center> |
| + | <p>We demonstrated that the bacteriophytochrome were functional by displaying their ability to bind biliverdin as well as by their presence in an SDS PAGE gel stained with Coomassie Blue. The gel for the bacteriophytochromes can be seen below,</p> | ||
| + | <center><img src="http://25.media.tumblr.com/tumblr_mazpn7P5iT1rg4kjpo1_500.jpg" style="width:80%"></center> | ||
| + | <p>The gel shows the expected bands for the bacteriophytochrome (Lanes 2, 5, 6, 7, 8) of approximately 80 kDA.We demonstrated that the bacteriophytochrome was functional and able to bind biliverdin in results below.</p> | ||
<hr> | <hr> | ||
| - | <center><a name="7"><h3>The Switch</h3></center> | + | <center><a name="7"><h3>The Switch</h3></a></center> |
| - | <p>Two of the BioBricks produced were ligated together to produce the light switch. We demonstrated that the switch was produced by inspecting a gel following ligation and then digestion. The gel can be seen below.</p> | + | <p>Two of the BioBricks produced (Heme Oxygenase and the Deinococcus Bacteriophytochrome) were ligated together to produce the light switch. We demonstrated that the switch was produced by inspecting a gel following ligation and then digestion. The gel can be seen below.</p> |
<center><img src="https://static.igem.org/mediawiki/2012/9/9f/GEL_ligation.jpeg"></center> | <center><img src="https://static.igem.org/mediawiki/2012/9/9f/GEL_ligation.jpeg"></center> | ||
| - | <p>Gel 1: We have run against a Heme Oxygenase standard (Lane 1). The gel contains digested fragments from our composite BioBrick (Heme Oxygenase and | + | <p>Gel 1: We have run against a Heme Oxygenase standard (Lane 1, 770 bp). The gel contains digested fragments from our composite BioBrick (Heme Oxygenase and Deinococcus). The upper band (Black Box) is the Heme Oxygenase with the bacteriophytochrome and the bottom band (blue) is the plasmid backbone.</p> |
<p>This provided the evidence that the product had been successfully ligated.</p> | <p>This provided the evidence that the product had been successfully ligated.</p> | ||
| - | <a name="gel"><h2>The | + | <a name="gel"><h2>The Gels</a></h2> |
| - | <p>An SDS page gel was run of the ligation products to observe if the heme oxygenase was able to produce biliverdin and then to determine if it was binding with the bacteriophytochrome. The biliverdin binds to a specific site in the bacteriophytochrome. As biliverdin is | + | <p>An SDS page gel was run of the ligation products to observe if the heme oxygenase was able to produce biliverdin and then to determine if it was binding with the bacteriophytochrome. The biliverdin binds to a specific site in the bacteriophytochrome. As biliverdin is fluorescent this coupling can be observed by irradiation with infrared (IR) light. </p> |
| - | <img src="https://static.igem.org/mediawiki/2012/d/ | + | <br><center><img src="https://static.igem.org/mediawiki/2012/d/d0/IRGELrationale.JPG" width=80%></center><br> |
| - | <p> | + | The SDS PAGE gels for the Switch Constructs confirmed that the genes were compatible and that we successfully produced a T7 controlled operon-like system. The annotated gel can be seen below,</p> |
| - | <img src="https://static.igem.org/mediawiki/2012/ | + | <center><img src="http://25.media.tumblr.com/tumblr_mazpn7P5iT1rg4kjpo1_500.jpg" style="width:80%"> |
| + | <br><p>The gel contains the following constructs:</p></center> | ||
| + | <center><table><tr><td><ul> | ||
| + | <li>The Protein Molecular Weight Marker</li> | ||
| + | <li>1C3C, the heme oxygenase and Deinococcus bacteriophytochrome ligation products (Lanes 2, 5, 6).</li> | ||
| + | <li>1C, the heme oxygenase (Lanes 3, 4)</li> | ||
| + | <li>4KE, Agro bacteriophytochrome (Lanes 7, 8).</li></ul></td></tr></table></center> | ||
| + | <br> | ||
| + | <p>We were able to show that the bacteriophytochrome was produced in the construct. Bands were observed at the same molecular weight as our bacteriophytochrome controls (4KE1 and 4KE3). We were able to visualise useing Coomassie Blue. Heme oxygenase was also produced in a low yield. We then determined that successfully the switch with heme oxygenase providing biliverdin for the bacteriophytochrome. This was observed under IR light to take advantage of biliverdin's fluorescence.</p> | ||
| + | <br><center><img src="https://static.igem.org/mediawiki/2012/8/83/Gel_Comparison.JPG" width=90%></center> | ||
| + | <p>The image contains four different SDS PAGE gels. The 2 on the left have been visualised using infrared light and the two on the right are the same gel stained with Coomassie Blue. This shows that many bands visible after Coomassie Blue staining are still not visible in the presence of IR light. For the band to be visualised a fluorophore is required. The top two gels act as a control as they contain no bacteriophytochrome, these are the T7 regulated Heme Oxygenase BioBricks we produced. This demonstrates that there are no native proteins, in a high concentration, that will cause false positives when the bacteriophytochrome is introduced.</p> | ||
| + | <p>The bottom two gels contain the Heme Oxygenase-Deinococcus bacteriophytochrome construct (1C3C), Heme Oxygenase (1C), and Agrobacterium bacteriophytochrome (4KE). The boxed area has been used to show that on select bands in the Coomassie stained gel are fluorescent in the infrared image. The blue circled area indicated that the bacteriophytochrome is not fluorescent in its native state. The bands that are active in the Infrared gel are the composite part we produced. It is our functional switch. The infrared image provides the information that:</p> | ||
| + | <blockquote><ul><li>Heme Oxygenase produces the fluorescent molecule Biliverdin.</li> | ||
| + | <li>In our switch, Biliverdin binds specifically to the bacteriophytochrome in <i>E. coli</i>.</li> | ||
| + | <li>The bacteriophytochrome is the only molecule that can be visualised in the infrared spectrum. It requires biliverdin to be present in the cell</li></ul></blockquote> | ||
| + | <p>The infrared gel shows that the switch is capable of self assembly without an external source of biliverdin. This is seen in the infrared gel as only the Switch's bacteriophytochromes can be visualised.</p> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2012/8/83/BV_gradient.png" width=80%></center> | ||
| + | <p>The gel contains: 4K1 (Agrobacterium T7) no biliverdin (BV), 4K1-25 uM BV, 4K1-50 uM BV, 4K3-no biliverdin, 4K3-25 uM BV, 4K3-50 uM BV, 1C3C-3 (Heme Oxygenase + Deino) no BV, 1C3C-3 25 uM BV, 1C3C-3 50 uM BV, 1C3C-6 no BV, 1C3C-7 no BV.</p> | ||
| + | <p>The gel shows that with an increasing gradient of biliverdin supplied to the cell there is no change in the intensity of the band in the infrared spectrum. Therefore we determined that our heme oxygenase part was able to supply the cell with enough biliverdin to saturate the bacteirophytochrome. </p> | ||
| + | <br> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2012/e/e2/Light_switch.png" width=50%></center> | ||
| + | <p>The spectrum of the cleared cell lysate of the Heme Oxygenase-Deinococcus Bacteriophytochrome expression shows that we have successfully assembled the light switch and in the Far-Red form. The peaks at 700 and 751 nm correlate with the literature values for the Far Red form Deinococcus bacteriophytochrome (Wagner et al. THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 283, NO. 18, pp. 12212–12226, May 2, 2008). We have placed underneath the spectrum the shape of the different peaks. We had no access to a light source to switch it back to the red form (760 nm) and so could not convert it back to the Red form.</p><hr> | ||
| + | <center><b><a name="switch"><h1>Reversing the Switch</h1></a></b></center> | ||
| + | <h3>Following the regional Jamboree we were able to show that the light switch was functional and was able to rapidly and reversible convert between the red and far red form.</h3></b></p><br> | ||
| + | <p>A dual beam UV-Vis spectrophotometer was used to provide a constant reference to an uninduced cell line's lysate, which experienced the same irradiation. The Agrobacterium bacteriophytochrome (4K) was used due to greater yield of protein. The induced cell lysate with no biliverdin. After this spectrum was obtained, the induced cell line with no ALA added was used as a reference. The spectrum for the bacteriophytochrome was collected in its native state (no treatment) and showed that it was assembled in the red form (yellow trace). Upon irradiation with 680 nm red light, it was able to be converted to the far red form (blue trace). This was seen in the spectrum with an increase in absorbance at 750 nm and decrease at 650 nm. We were able to get the switch to convert back from the far-red form to the red form by irradiation with 680 nm light (blue trace) for one minute. This data can be seen in the spectra below. The lysates were stored in the dark at -20°C allowing for the bacteriophytochrome to relax back to the red form. After 3 days the spectra were collected and shown to be reproducible.</p> | ||
| + | <br><center><img src="https://static.igem.org/mediawiki/2012/3/37/4Kspectra.png"></center> | ||
| + | <center>The spectra show that the switch could successfully convert between the two spectral forms</center> | ||
| + | </body></html> | ||
Latest revision as of 10:11, 26 October 2012
Results and Characterisation
|
|
|
|---|---|
Heme Oxygenase |
Bacteriophytochromes |
Heme Oxygenase |
Bacteriophytochromes |
| Characterisation | |||
|---|---|---|---|
Heme Oxygenase |
Bacteriophytochromes |
Building The Switch |
Reversing The Switch |
Heme Oxygenase Results
We produced a Heme Oxygenase BioBrick that was codon optimized for E. coli. The Gibson assembly of the T7 promoter containing Heme Oxygenase was successful. The transformation was successful with numerous colonies grown using Chloramphenicol as the selecting agent. Six colonies were selected and then they were sequenced before digestion with EcoR1 and Spe1. The sequencing showed that all of the colonies contained the plasmid with a Heme oxygenase identical to the original protein sequence. The gel containing the digested Heme Oxygenase bearing plasmid can be seen in Figure 1.
 |
|
and the heme oxygenese gene (Green Box). We used a 1kb ladder (in increasing Kbp: 0.25, 0.5, 0.75, 1, 1.5, 2, 2.5, 3, 4, 5, 6, 8, 10). |
The lower band is the T7 Heme Oxygenase BioBrick (771 bp) which was expected. Similarly, the plasmid backbone was visible at the expected length (2070 bp). The similarity in intensity of the bands also demonstrates that the assembly and the digest were successful. The sequencing results for the six plasmids above can be seen below.
Heme Oxygenase Sequencing Results
The Gibson Assembly performed was transformed into 6 different cultures of competent cells. Three of theses were selected for sequencing using both forward and reverse primers. They were sequenced using the forward and reverse primers for the BioBricks. We performed Blastx and Blastn pipelines (available here) to determine if there was a significant change in the protein sequence and to determine the identity of the plasmid. The sequences below are named to show: that it is heme oxygenase (1C), which transformation (4,5,6), and whether it is the forward or reverse primer (F or R).
| Sample | Identity | E-value | MaxID |
| 1C-6F | Heme Oxygenase (Synechocystic sp. PCC603) | 7e-176 | 99% |
| 1C-6R | Heme Oxygenase (Synechocystic sp. PCC603) | 1e-171 | 99% |
| 1C-4F | Heme Oxygenase (Synechocystic sp. PCC603) | 4e-176 | 99% |
| 1C-4R | Heme Oxygenase (Synechocystic sp. PCC603) | 6e-172 | 99% |
| 1C-5F | Heme Oxygenase (Synechocystic sp. PCC603) | 6e-172 | 99% |
|
The source of our gene was identified in the sequencing result which showed that the sequencing was accurate. We then compared to the original gBlock sequence and determined that the sequencing was accurate and confirmed the identity of the plasmid. The Blastn pipeline indicated that there was no significant change from the theoretical sequence. With this data we would assume that the protein would be functional and performed assays to determine if this was the case.
Characterisation of Heme Oxygenase
The T7 regulated Heme Oxygenase produced was characterised to determine its functionality. BL21 E. coli was transformed with the plasmid, selected for using chloramphenicol, and a culture was inoculated. The culture was then induced with ALA (d-aminolevulinic acid) for the heme pathway and IPTG to promote protein production. They were incubated overnight and the cells were spun down. We observed a functional Heme Oxygenase with the cells appeared a vibrant green after induction by ALA and IPTG. We observed this as well in our assembled switch. The image below demonstrates the green pigment produced by the cells compared to uninduced Heme Oxygenase and the bacteriophytochrome.
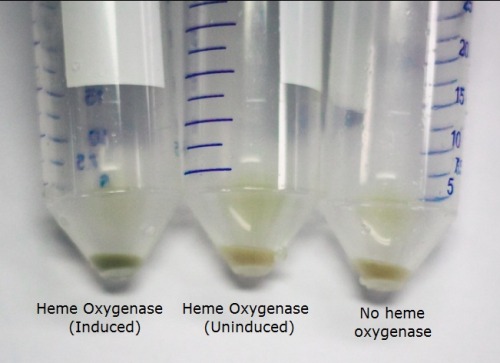
The cells were lysed and the protein extract run on an SDS page gel. The gel showed the protein bands near the expected size (27 kDa). This, along with our pigmentation, showed that the BioBrick was producing the functional protein.

Bacteriophytochromes Results
Like the Heme Oxygenase, the bacteriophytochromes from Deinococcus radiodurans and Agrobacterium tumefaciens were codon optimised for use in E. Coli. We did not use genomic DNA or previous Macquarie University team DNA at any stage, everything was performed with synthetic DNA we designed. The identity of the plasmid was determined by sequencing and by digestion. The bacteriophytochromes were characterised by demonstrating their ability to bind biliverdin and by their characteristic absorption profile.

| The restriction enzymes used in this digestion were, |
|
| We used a 1kb ladder (in increasing Kbp: 0.25, 0.5, 0.75, 1, 1.5, 2, 2.5, 3, 4, 5, 6, 8, 10). |
The digest above shows only shows one band for the different digests performed on the Agro T7 plasmid. The length of the vector and the bacteriophytochrome are similar and make it difficult to resolve the two bands. Only the digested plasmid can be seen which indicates that the bacteriophytochrome component was assembled correctly. Based on the length in base pairs of the fragments (2.25 kbp) we determined that assembly was successful. The first Agro with no T7 digest (asterixed band) shows incomplete digestion of a BioBrick plasmid. This enacted as a control and indicated the size of the plasmid.
Bacteriophytochrome Sequencing
Plasmids extracted from the different transformations were sequenced.
| Sequence | Expected Identity | Identity | E value | Max ID |
| F3CE1 | Deinococcus radiodurans Bacteriophytochrome | Chain A, Crystal Structure Of A Monomeric Infrared Fluorescent Deinococcus Radiodurans Bacteriophytochrome Chromophore Binding Domain | 3.00E-160 | 99% |
| F3CE2 | Deinococcus radiodurans Bacteriophytochrome | Chain A, Crystal Structure Of A Monomeric Infrared Fluorescent Deinococcus Radiodurans Bacteriophytochrome Chromophore Binding Domain | 2.00E-161 | 99% |
| F3CE3 | Deinococcus radiodurans Bacteriophytochrome | Chain A, Crystal Structure Of A Monomeric Infrared Fluorescent Deinococcus Radiodurans Bacteriophytochrome Chromophore Binding Domain | 8.00E-163 | 99% |
| R4KE2 | Agrobacterium tumefaciens Bacteriophytochrome | bacteriophytochrome protein [Agrobacterium tumefaciens str. C58] | 0 | 99% |
| R5CE1 | Agrobacterium tumefaciens Bacteriophytochrome | bacteriophytochrome protein [Agrobacterium tumefaciens str. C58] | 0 | 99% |
| R5CE3 | Agrobacterium tumefaciens Bacteriophytochrome | bacteriophytochrome protein [Agrobacterium tumefaciens str. C58] | 0 | 99% |
|
The The Blastx pipeline showed that there was an identical match to the orginal source. This shows that the Gibson Assembly reactions were successful and we expect the protein to be functional. Blastn were run to determine the deviance from the theoretical sequence. The Blastn searches produced indicated that the sequences were nearly identical. The changed bases were examined on the sequencing output and determined to be possible misreads.
Bacteriophytochrome Characterisation
We demonstrated that the bacteriophytochrome were functional by displaying their ability to bind biliverdin as well as by their presence in an SDS PAGE gel stained with Coomassie Blue. The gel for the bacteriophytochromes can be seen below,
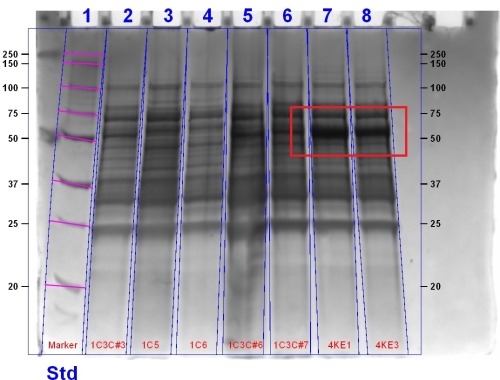
The gel shows the expected bands for the bacteriophytochrome (Lanes 2, 5, 6, 7, 8) of approximately 80 kDA.We demonstrated that the bacteriophytochrome was functional and able to bind biliverdin in results below.
The Switch
Two of the BioBricks produced (Heme Oxygenase and the Deinococcus Bacteriophytochrome) were ligated together to produce the light switch. We demonstrated that the switch was produced by inspecting a gel following ligation and then digestion. The gel can be seen below.

Gel 1: We have run against a Heme Oxygenase standard (Lane 1, 770 bp). The gel contains digested fragments from our composite BioBrick (Heme Oxygenase and Deinococcus). The upper band (Black Box) is the Heme Oxygenase with the bacteriophytochrome and the bottom band (blue) is the plasmid backbone.
This provided the evidence that the product had been successfully ligated.
The Gels
An SDS page gel was run of the ligation products to observe if the heme oxygenase was able to produce biliverdin and then to determine if it was binding with the bacteriophytochrome. The biliverdin binds to a specific site in the bacteriophytochrome. As biliverdin is fluorescent this coupling can be observed by irradiation with infrared (IR) light.

The SDS PAGE gels for the Switch Constructs confirmed that the genes were compatible and that we successfully produced a T7 controlled operon-like system. The annotated gel can be seen below,

The gel contains the following constructs:
|
We were able to show that the bacteriophytochrome was produced in the construct. Bands were observed at the same molecular weight as our bacteriophytochrome controls (4KE1 and 4KE3). We were able to visualise useing Coomassie Blue. Heme oxygenase was also produced in a low yield. We then determined that successfully the switch with heme oxygenase providing biliverdin for the bacteriophytochrome. This was observed under IR light to take advantage of biliverdin's fluorescence.

The image contains four different SDS PAGE gels. The 2 on the left have been visualised using infrared light and the two on the right are the same gel stained with Coomassie Blue. This shows that many bands visible after Coomassie Blue staining are still not visible in the presence of IR light. For the band to be visualised a fluorophore is required. The top two gels act as a control as they contain no bacteriophytochrome, these are the T7 regulated Heme Oxygenase BioBricks we produced. This demonstrates that there are no native proteins, in a high concentration, that will cause false positives when the bacteriophytochrome is introduced.
The bottom two gels contain the Heme Oxygenase-Deinococcus bacteriophytochrome construct (1C3C), Heme Oxygenase (1C), and Agrobacterium bacteriophytochrome (4KE). The boxed area has been used to show that on select bands in the Coomassie stained gel are fluorescent in the infrared image. The blue circled area indicated that the bacteriophytochrome is not fluorescent in its native state. The bands that are active in the Infrared gel are the composite part we produced. It is our functional switch. The infrared image provides the information that:
- Heme Oxygenase produces the fluorescent molecule Biliverdin.
- In our switch, Biliverdin binds specifically to the bacteriophytochrome in E. coli.
- The bacteriophytochrome is the only molecule that can be visualised in the infrared spectrum. It requires biliverdin to be present in the cell
The infrared gel shows that the switch is capable of self assembly without an external source of biliverdin. This is seen in the infrared gel as only the Switch's bacteriophytochromes can be visualised.

The gel contains: 4K1 (Agrobacterium T7) no biliverdin (BV), 4K1-25 uM BV, 4K1-50 uM BV, 4K3-no biliverdin, 4K3-25 uM BV, 4K3-50 uM BV, 1C3C-3 (Heme Oxygenase + Deino) no BV, 1C3C-3 25 uM BV, 1C3C-3 50 uM BV, 1C3C-6 no BV, 1C3C-7 no BV.
The gel shows that with an increasing gradient of biliverdin supplied to the cell there is no change in the intensity of the band in the infrared spectrum. Therefore we determined that our heme oxygenase part was able to supply the cell with enough biliverdin to saturate the bacteirophytochrome.

The spectrum of the cleared cell lysate of the Heme Oxygenase-Deinococcus Bacteriophytochrome expression shows that we have successfully assembled the light switch and in the Far-Red form. The peaks at 700 and 751 nm correlate with the literature values for the Far Red form Deinococcus bacteriophytochrome (Wagner et al. THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 283, NO. 18, pp. 12212–12226, May 2, 2008). We have placed underneath the spectrum the shape of the different peaks. We had no access to a light source to switch it back to the red form (760 nm) and so could not convert it back to the Red form.
Reversing the Switch
Following the regional Jamboree we were able to show that the light switch was functional and was able to rapidly and reversible convert between the red and far red form.
A dual beam UV-Vis spectrophotometer was used to provide a constant reference to an uninduced cell line's lysate, which experienced the same irradiation. The Agrobacterium bacteriophytochrome (4K) was used due to greater yield of protein. The induced cell lysate with no biliverdin. After this spectrum was obtained, the induced cell line with no ALA added was used as a reference. The spectrum for the bacteriophytochrome was collected in its native state (no treatment) and showed that it was assembled in the red form (yellow trace). Upon irradiation with 680 nm red light, it was able to be converted to the far red form (blue trace). This was seen in the spectrum with an increase in absorbance at 750 nm and decrease at 650 nm. We were able to get the switch to convert back from the far-red form to the red form by irradiation with 680 nm light (blue trace) for one minute. This data can be seen in the spectra below. The lysates were stored in the dark at -20°C allowing for the bacteriophytochrome to relax back to the red form. After 3 days the spectra were collected and shown to be reproducible.

 "
"




