Team:LMU-Munich/Spore Coat Proteins/mainresult
From 2012.igem.org
Franzi.Duerr (Talk | contribs) |
|||
| (11 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
===GFP as a Proof of Principle=== | ===GFP as a Proof of Principle=== | ||
| - | <p align="justify">We | + | <p align="justify">We designed the first GFP-'''Sporo'''bead to evaluate our system and compare different constructs. |
| - | + | <br> | |
| + | This short time lapse video shows the formation of a spore.</p> | ||
| + | <br> | ||
| + | {| width="100%" | ||
| + | | | ||
| + | <html><div align="center"> | ||
| + | <iframe width="315" height="315" src="http://www.youtube.com/embed/fXhUsB4dTIs" frameborder="0" allowfullscreen></iframe></div> | ||
| + | </html> | ||
| + | |} | ||
| + | <br> | ||
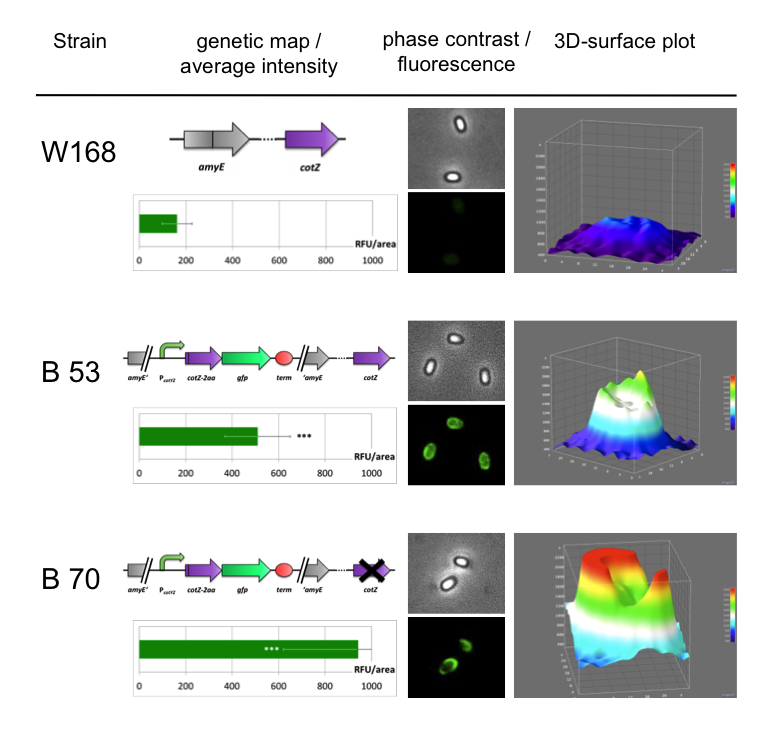
| + | <p align="justify">To ensure that our '''Sporo'''beads show fluorescence on their edge and higher than in wildtype spores, we analysed the microscopy pictures. The following graph (Fig. 6) shows these microscopy pictures and analysis of the strongest of the five constructs integrated into wildtype strain (B53) and the deletion strain (B70).</p> | ||
| + | <br> | ||
{| style="color:black;" cellpadding="3" width="100%" cellspacing="0" border="0" align="center" style="text-align:left;" | {| style="color:black;" cellpadding="3" width="100%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| style="width: 70%;background-color: #EBFCE4;" | | | style="width: 70%;background-color: #EBFCE4;" | | ||
{| | {| | ||
| - | |[[File:Fluorescence of Sporobeads.png|610px|center]] | + | |[[File:Fluorescence of Sporobeads.png|610px|center|link=]] |
|} | |} | ||
|- | |- | ||
| Line 23: | Line 34: | ||
{| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;" | {| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
|style="width: 70%;background-color: #EBFCE4;" | | |style="width: 70%;background-color: #EBFCE4;" | | ||
| - | <font color="#000000"; size="2">Fig. 6: Result of fluorescence evaluation of the three strains W168, B53 and B70.</font> | + | <font color="#000000"; size="2">Fig. 6: Result of fluorescence evaluation of the three strains W168, B53 and B70. The bar charts show the average fluorescence intensity over 100-200 spores. For image analysis we measured the fluorescence intensity of an area of 750 pixel per spore by using ImageJ and evaluated the results with the statistical software R. The 3D graphs illustrate in one spore the distribution of fluorescence intensity across the spore surface.</font> |
|} | |} | ||
|} | |} | ||
| - | <p align="justify">As shown in Fig. 6, the wild type spore has hardly any fluorescence, whereas both''' Sporo'''beads with the integrated construct pSB<sub>''Bs''</sub>1C-P<sub>''cotYZ''</sub>-''cotZ''<sub>-2aa</sub>-''gfp''-terminator give a distinct fluorescence signal around the edge of the spore. Furthermore, it demonstrates that strain B 70 has the highest fluorescence intensity. For more detailed information look at our | + | <p align="justify">As shown in Fig. 6, the wild type spore has hardly any fluorescence, whereas both''' Sporo'''beads with the integrated construct pSB<sub>''Bs''</sub>1C-P<sub>''cotYZ''</sub>-''cotZ''<sub>-2aa</sub>-''gfp''-terminator give a distinct fluorescence signal around the edge of the spore. Furthermore, it demonstrates that strain B 70 has the highest fluorescence intensity. For more detailed information have later look at our DATA page</p> |
<p align="justify">In summary we successfully developed functional '''Sporo'''beads that are capable of displaying any protein of choice on the surface of modified ''B. subtilis'' endospores.</p> | <p align="justify">In summary we successfully developed functional '''Sporo'''beads that are capable of displaying any protein of choice on the surface of modified ''B. subtilis'' endospores.</p> | ||
| - | |||
| - | |||
Latest revision as of 16:48, 26 October 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]


GFP as a Proof of Principle
We designed the first GFP-Sporobead to evaluate our system and compare different constructs.
This short time lapse video shows the formation of a spore.
|
|
To ensure that our Sporobeads show fluorescence on their edge and higher than in wildtype spores, we analysed the microscopy pictures. The following graph (Fig. 6) shows these microscopy pictures and analysis of the strongest of the five constructs integrated into wildtype strain (B53) and the deletion strain (B70).
| |
|
As shown in Fig. 6, the wild type spore has hardly any fluorescence, whereas both Sporobeads with the integrated construct pSBBs1C-PcotYZ-cotZ-2aa-gfp-terminator give a distinct fluorescence signal around the edge of the spore. Furthermore, it demonstrates that strain B 70 has the highest fluorescence intensity. For more detailed information have later look at our DATA page
In summary we successfully developed functional Sporobeads that are capable of displaying any protein of choice on the surface of modified B. subtilis endospores.
 "
"