Team:LMU-Munich/Spore Coat Proteins
From 2012.igem.org
Franzi.Duerr (Talk | contribs) |
|||
| (24 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
<!-- Include the next line at the beginning of every page --> | <!-- Include the next line at the beginning of every page --> | ||
{{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_eppis.resized.jpg|3}} | {{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_eppis.resized.jpg|3}} | ||
| + | [[File:LMU Glow Spore2 cutII.jpg|620px|link=]] | ||
| - | + | [[File:SporeCoat.png|100px|right|link=]] | |
| - | + | ||
| - | [[File:SporeCoat.png|100px|right|link= | + | |
| - | + | ||
| Line 12: | Line 10: | ||
=='''Sporo'''beads - What Protein Do ''You'' Want to Display?== | =='''Sporo'''beads - What Protein Do ''You'' Want to Display?== | ||
<br> | <br> | ||
| + | <p align="justify">'''Sporo'''beads are the first generation of ''Bacillus subtilis'' endospores displaying a protein of our choice on their outermost layer, the spore crust. In future '''Sporo'''beads could serve as a platform for protein display and thus be used for numerous versatile [https://2012.igem.org/Team:LMU-Munich/Application applications]. Our goal was to show that ''B. subtilis'' spores have the ability to do so. As a [https://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins/result proof of principle] we successfully fused GFP to the spore crust and obtained fluorescence with microscopy.</p> | ||
| + | |||
| + | |||
<div class="box"> | <div class="box"> | ||
| - | === | + | ===Scientific Background=== |
{| "width=100%" style="text-align:center;" style="align:right"| | {| "width=100%" style="text-align:center;" style="align:right"| | ||
| - | |<p align="justify">Introduction to ''B. subtilis'' spores and their | + | |<p align="justify">Introduction to ''B. subtilis'' spores and their use in our project</p> |
|[[File:Imamura, 2011 & McKenney, 2010.png|200px|right|link=Team:LMU-Munich/Spore_Coat_Proteins/Background]] | |[[File:Imamura, 2011 & McKenney, 2010.png|200px|right|link=Team:LMU-Munich/Spore_Coat_Proteins/Background]] | ||
|- | |- | ||
| Line 23: | Line 24: | ||
<div class="box"> | <div class="box"> | ||
| - | === | + | ===Cloning Strategy=== |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{| "width=100%" style="text-align:center;" style="align:right"| | {| "width=100%" style="text-align:center;" style="align:right"| | ||
| - | |<p align="justify"> | + | |<p align="justify">Cloning strategy to create different variants of our ''' Sporo'''beads</p> |
| - | |[[File: | + | |[[File:Final construct.png|200px|link=Team:LMU-Munich/Spore_Coat_Proteins/cloning]] |
|- | |- | ||
| - | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Spore_Coat_Proteins/ | + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Spore_Coat_Proteins/cloning]] |
|} | |} | ||
</div> | </div> | ||
<div class="box"> | <div class="box"> | ||
| - | === | + | ===GFP as a Proof of Principle=== |
{| "width=100%" style="text-align:center;" style="align:right"| | {| "width=100%" style="text-align:center;" style="align:right"| | ||
| - | |<p align="justify"> | + | |<p align="justify">Main results of the various constructs that were created to find the best one!</p> |
| - | |[[File: | + | |[[File:LMU Firstspore.jpg|200px|right|link=Team:LMU-Munich/Spore_Coat_Proteins/result]] |
|- | |- | ||
| - | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Spore_Coat_Proteins/ | + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Spore_Coat_Proteins/result]] |
|} | |} | ||
</div> | </div> | ||
<div class="box"> | <div class="box"> | ||
| - | === | + | ===Laccases as functional enzymes=== |
{| "width=100%" style="text-align:center;" style="align:right"| | {| "width=100%" style="text-align:center;" style="align:right"| | ||
| - | |<p align="justify"> | + | |<p align="justify">Creation of functional Laccase-''' Sporo'''beads</p> |
| - | |[[File: | + | |[[File:Final construct.png|200px|link=Team:LMU-Munich/Spore_Coat_Proteins/laccases]] |
|- | |- | ||
| - | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Spore_Coat_Proteins/ | + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Spore_Coat_Proteins/laccases]] |
|} | |} | ||
</div> | </div> | ||
<div class="box"> | <div class="box"> | ||
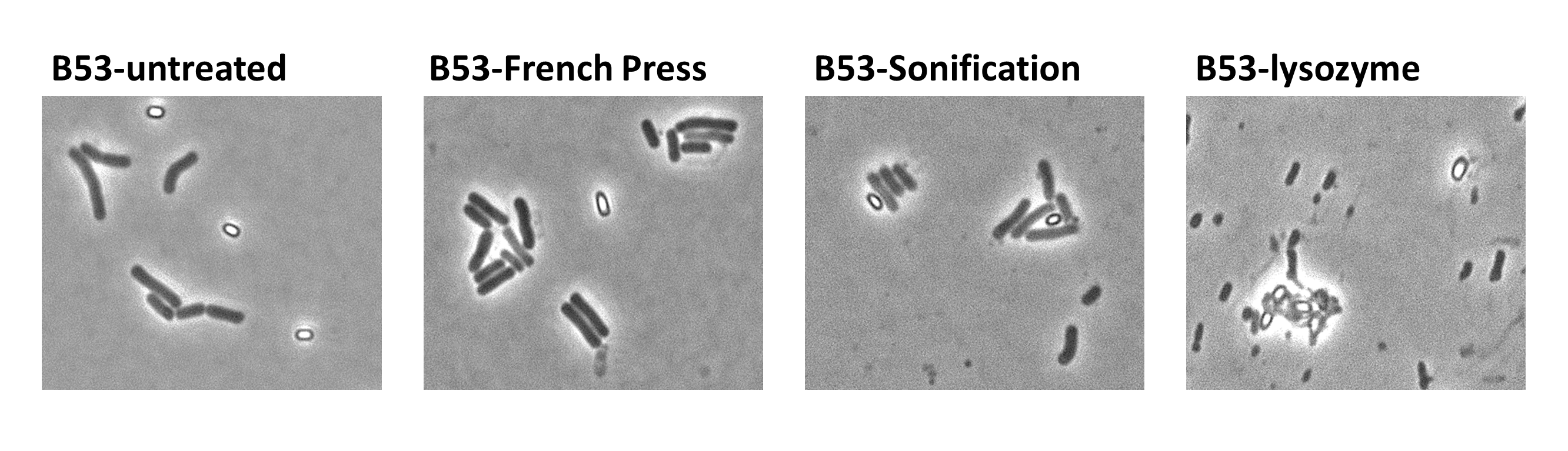
| - | ===Purification | + | ===Purification Methods=== |
{| "width=100%" style="text-align:center;" style="align:right"| | {| "width=100%" style="text-align:center;" style="align:right"| | ||
|<p align="justify">Description of the different purification methods of the spores</p> | |<p align="justify">Description of the different purification methods of the spores</p> | ||
| Line 71: | Line 62: | ||
|} | |} | ||
</div> | </div> | ||
| - | + | <br> | |
| - | + | <br> | |
| - | < | + | <br> |
| - | + | <br> | |
| - | < | + | <br> |
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<div class="box"> | <div class="box"> | ||
====Project Navigation==== | ====Project Navigation==== | ||
| Line 115: | Line 81: | ||
|} | |} | ||
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Latest revision as of 13:44, 14 November 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

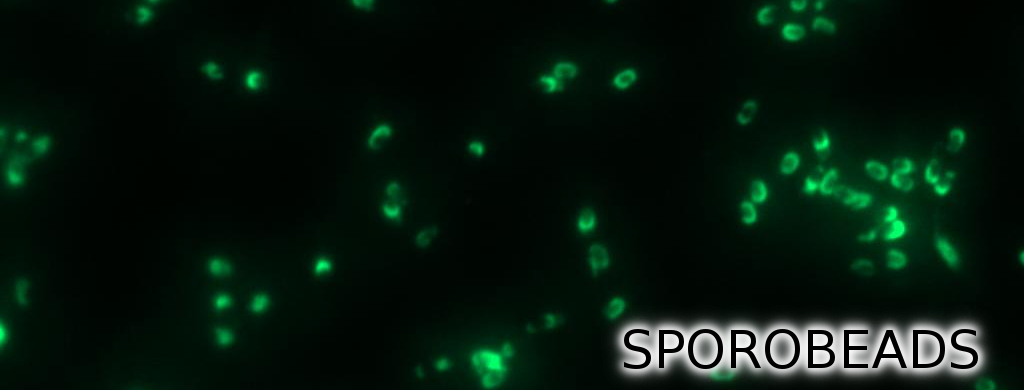

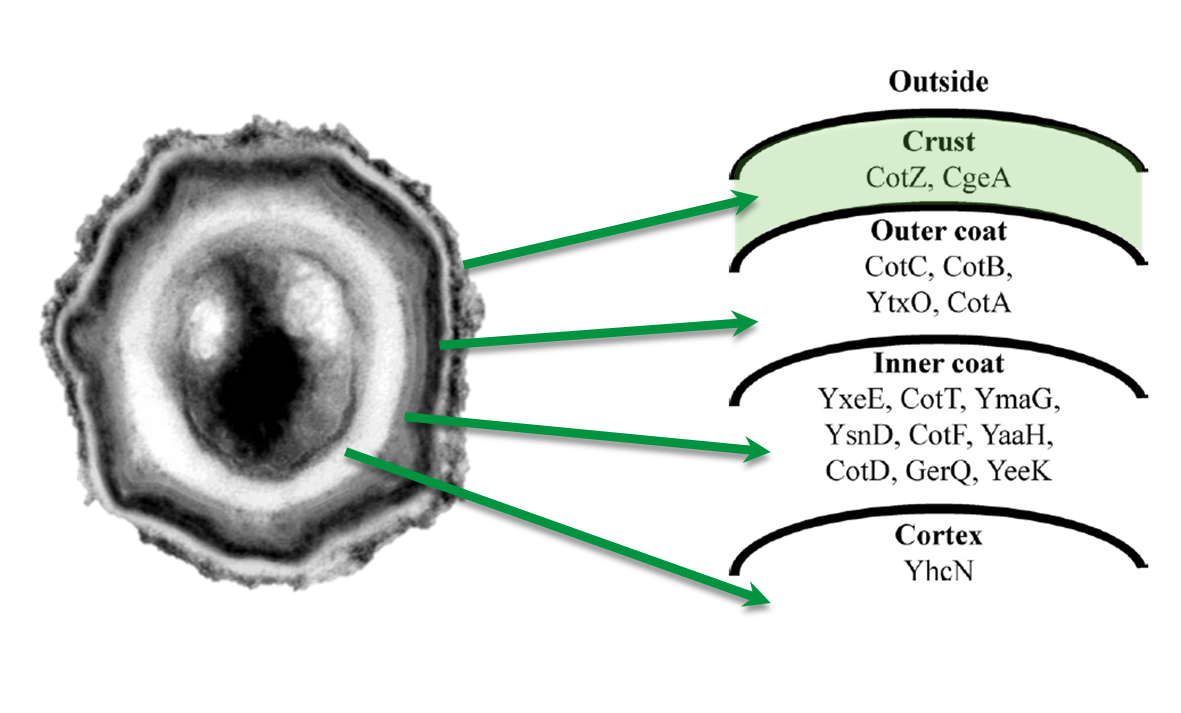
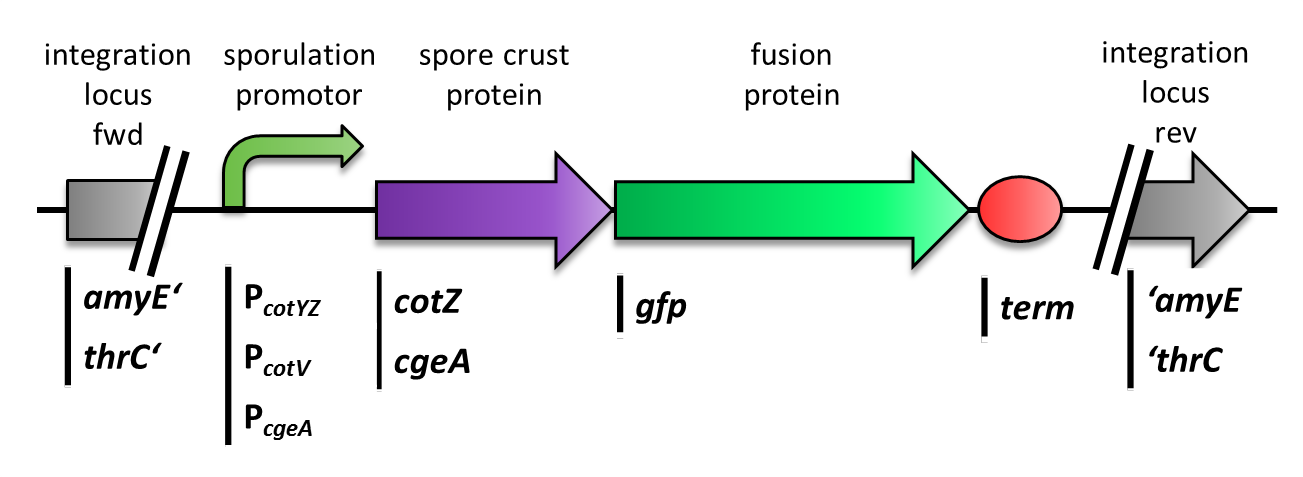
Sporobeads - What Protein Do You Want to Display?
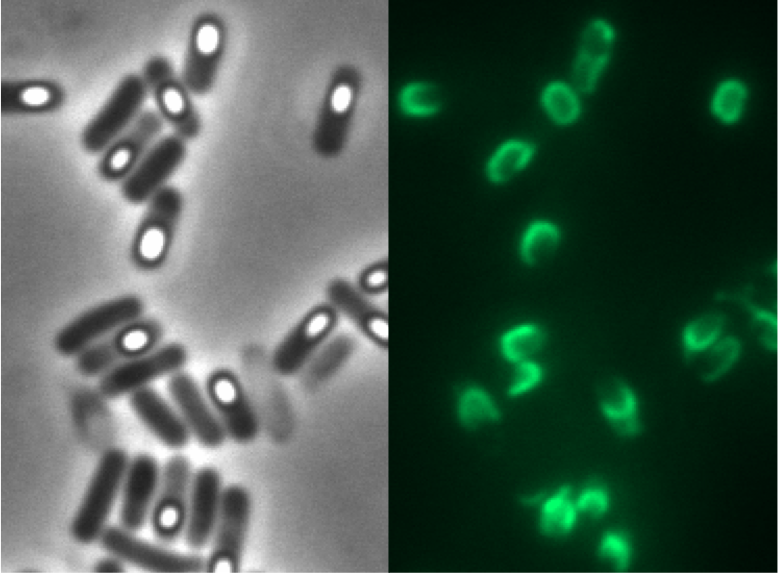
Sporobeads are the first generation of Bacillus subtilis endospores displaying a protein of our choice on their outermost layer, the spore crust. In future Sporobeads could serve as a platform for protein display and thus be used for numerous versatile applications. Our goal was to show that B. subtilis spores have the ability to do so. As a proof of principle we successfully fused GFP to the spore crust and obtained fluorescence with microscopy.
GFP as a Proof of Principle
Main results of the various constructs that were created to find the best one! | |

| 
| 
| 
|
| Bacillus Intro | Bacillus BioBrickBox | Sporobeads | Germination STOP |
 "
"