|
|
| Line 63: |
Line 63: |
| | | | |
| | <div class="box"> | | <div class="box"> |
| - | ===Purification methods=== | + | ===Purification Methods=== |
| | {| "width=100%" style="text-align:center;" style="align:right"| | | {| "width=100%" style="text-align:center;" style="align:right"| |
| | |<p align="justify">Description of the different purification methods of the spores</p> | | |<p align="justify">Description of the different purification methods of the spores</p> |
| Line 71: |
Line 71: |
| | |} | | |} |
| | </div> | | </div> |
| - |
| |
| - |
| |
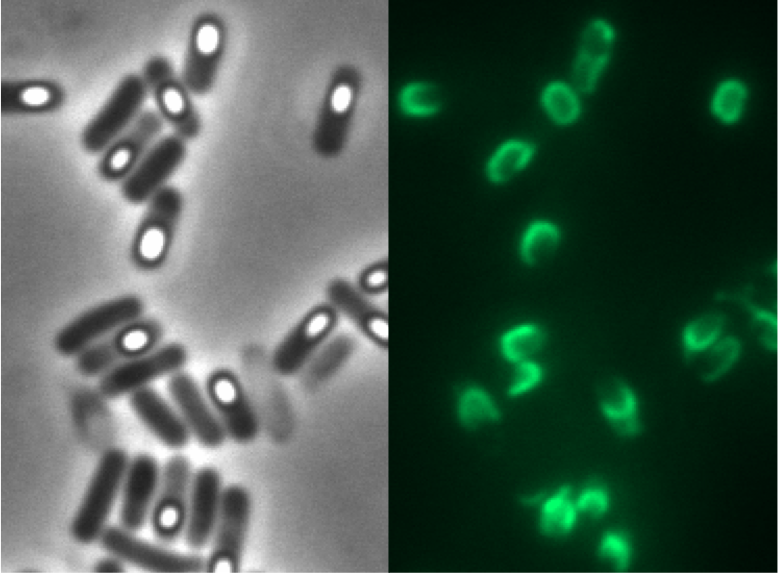
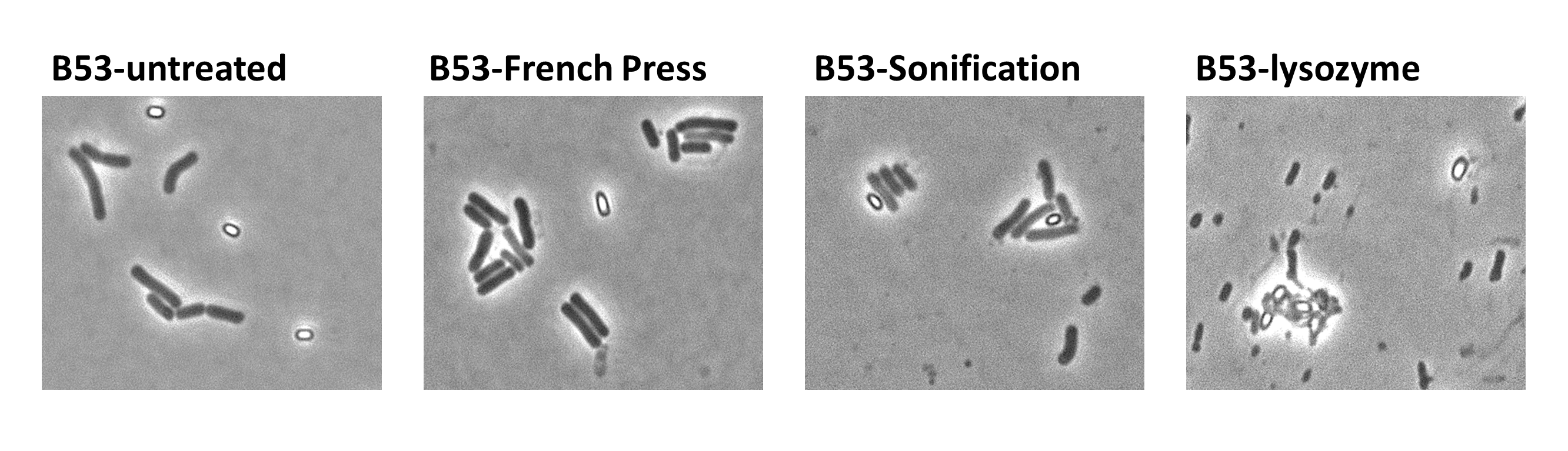
| - | <p align="justify">Since there were still some vegetative cells left after 24 hours of growth in Difco Sporulation Medium, we wanted to purify the '''Sporo'''beads. We tried three different methods for this approach: treatment with French Press, sonification and lysozyme. By means of the microscopy results we were able to conclude that lysozyme treatment was the only successful method (see [https://2012.igem.org/Team:LMU-Munich/Data/Sporepurification data]). Additionally, this treatment did not harm the crust fusion proteins as green fluorescence was still detectable afterwards (see [https://2012.igem.org/Team:LMU-Munich/Data/Sporepurification data] for details).</p>
| |
| - |
| |
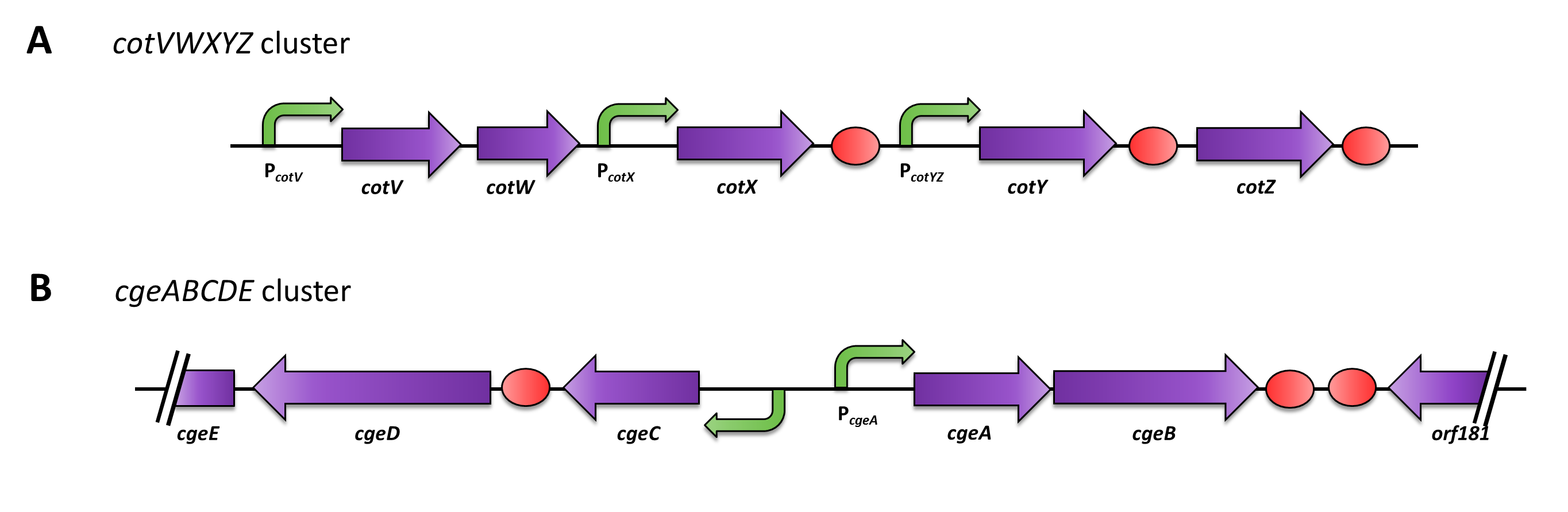
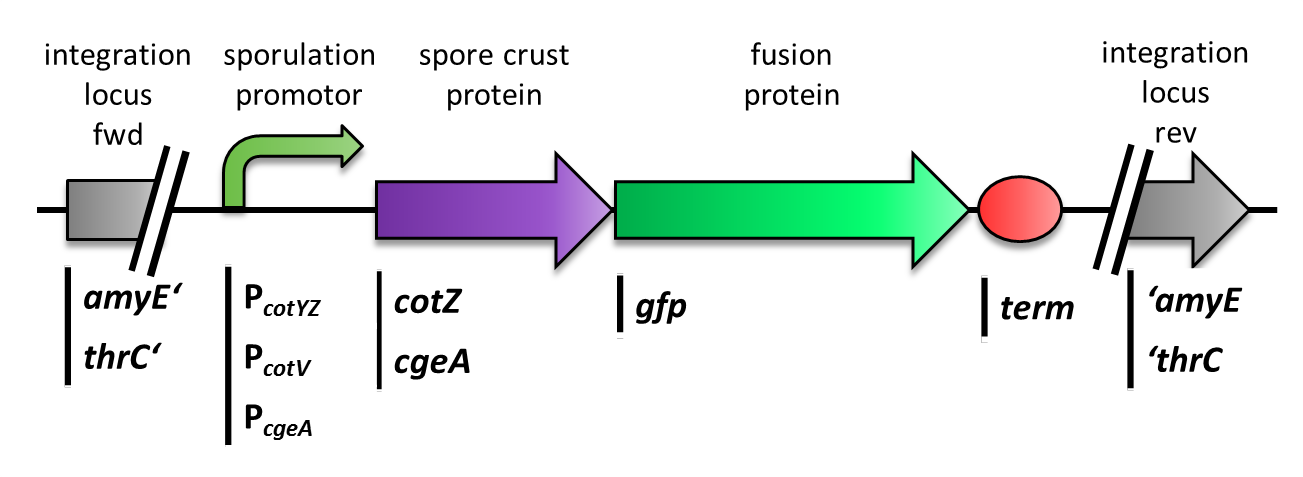
| - | <p align="justify">Moreover, we wondered if clean deletions of the native spore crust genes would show any difference in fusion protein expression in our '''Sporo'''beads. Thus, we deleted the native ''cotZ'' and ''cgeA'' using the pMAD based gene deletion strategy described by [http://www.ncbi.nlm.nih.gov/pubmedterm=New%20Vector%20for%20Efficient%20Allelic%20Replacement%20in%20Naturally%20Nontransformable%2C%20Low-GC-Content%2C%20Gram-Positive%20Bacteria Arnaud ''et al''., 2004].</p>
| |
| - |
| |
| - |
| |
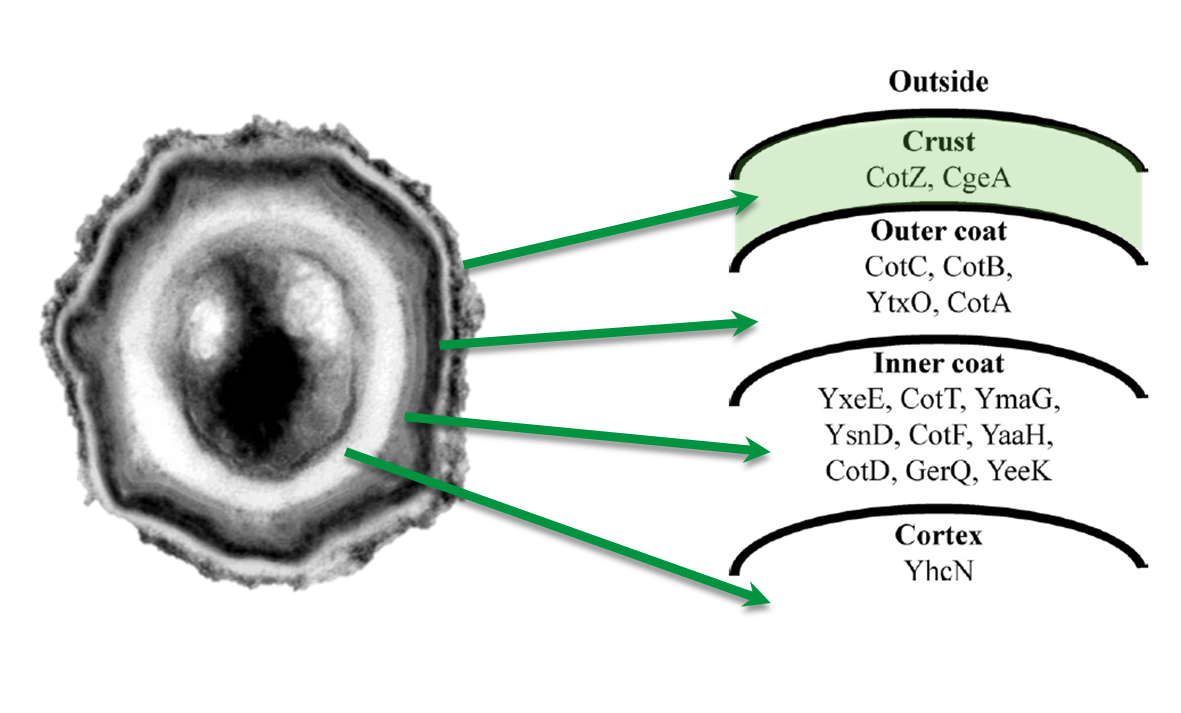
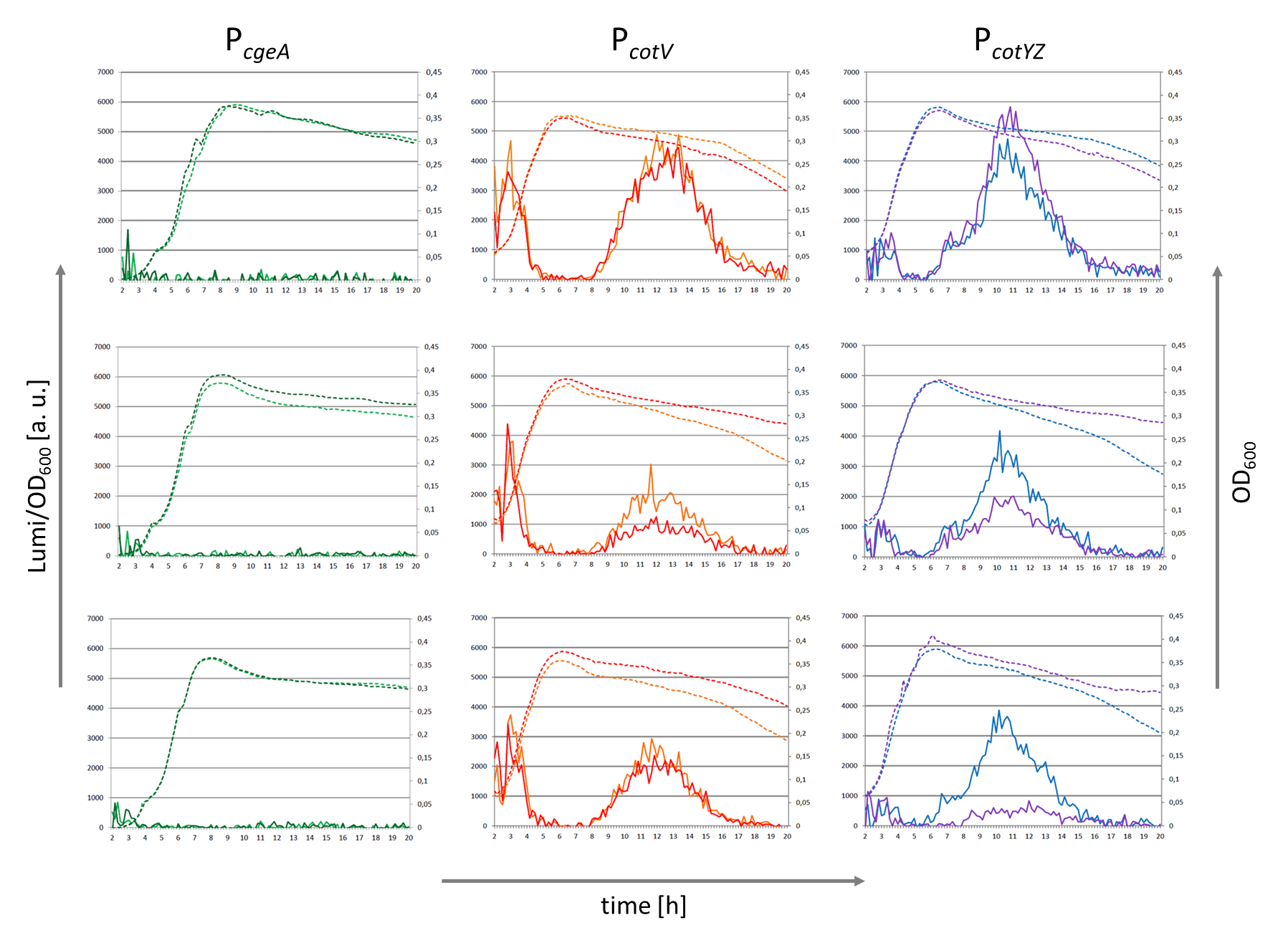
| - | <p align="justify">Because of the low but distinct fluorescence of wild type spores, we measured and compared the fluorescence intensity of 100 spores per construct (see [https://2012.igem.org/Team:LMU-Munich/Data/gfp_spore data]). We obtained significant differences between wild type spores and all of our '''Sporo'''beads (see [https://2012.igem.org/Team:LMU-Munich/Data/gfp_spore data]). The intensity bar charts in Fig. 6 show the fluorescence intensity, while the 3D graphs illustrate the distribution of fluorescence intensity across the spore surface. This correlates with the localization of our fusion proteins in the crust. For image analysis we measured the fluorescence intensity of an area of 750 pixel per spore by using ImageJ and evaluated the results with the statistical software R.
| |
| - | The following graph (Fig. 6) shows the results of microscopy and ImageJ analysis of the strongest construct integrated into wildtype W168 (B53) and the deletion strain B 49 (B70).</p>
| |
| - |
| |
| - |
| |
| - | {| style="color:black;" cellpadding="3" width="100%" cellspacing="0" border="0" align="center" style="text-align:left;"
| |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| |
| - | {|
| |
| - | |[[File:Fluorescence of Sporobeads.png|610px|center]]
| |
| - | |}
| |
| - | |-
| |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| |
| - | {| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;"
| |
| - | |style="width: 70%;background-color: #EBFCE4;" |
| |
| - | <font color="#000000"; size="2">Fig. 6: Result of fluorescence evaluation of the three strains W168, B53 and B70.</font>
| |
| - | |}
| |
| - | |}
| |
| - |
| |
| - |
| |
| - | <p align="justify">As shown in Fig. 6, the wild type spore has hardly any fluorescence, whereas both''' Sporo'''beads with the integrated construct pSB<sub>''Bs''</sub>1C-P<sub>''cotYZ''</sub>-''cotZ''<sub>-2aa</sub>-''gfp''-terminator give a distinct fluorescence signal around the edge of the spore. Furthermore, it demonstrates that strain B 70 has the highest fluorescence intensity.</p>
| |
| - |
| |
| - | <p align="justify">In summary we successfully developed functional sporobeads that are capable of displaying any protein of choice on the surface of modified ''B. subtilis'' endospores.</p>
| |
| | | | |
| | | | |
| Line 115: |
Line 87: |
| | |} | | |} |
| | </div> | | </div> |
| - |
| |
| - |
| |
| - | [[File:Spore crust proteins cycle.jpg|600px|right]]
| |
| - | {| style="color:black;" cellpadding="3" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;"
| |
| - | | style="width: 100%;background-color: #FFFFFF;" |
| |
| - | |-
| |
| - | | style="width: 70%;background-color: #FFFFFF;" |
| |
| - | {| style="color:black;" cellpadding="0" width="70%" cellspacing="0" border="0" align="center" style="text-align:center;"
| |
| - | |style="width: 70%;background-color: #FFFFFF;" |
| |
| - | <font color="#000000"; size="2">Fig. 3: The '''Bead'''zillus cycle</font>
| |
| - | |}
| |
| - | |}
| |
| - |
| |
| | | | |
| | | | |
| | <!-- Include the next line at the end of every page --> | | <!-- Include the next line at the end of every page --> |
| | {{:Team:LMU-Munich/Templates/Page Footer}} | | {{:Team:LMU-Munich/Templates/Page Footer}} |






 "
"